news
Publication: paper on an anti-polyglutamine oligomeric chaperone studies published!
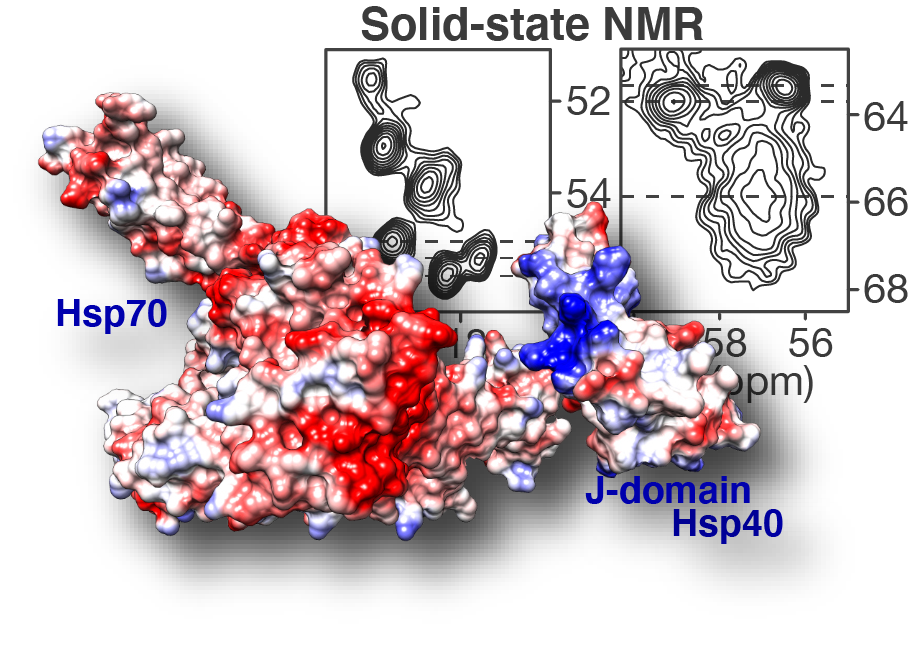
Congratulations to our postdoc Dr. Irina Matlahov and our collaborators in the group of Prof. Lukasz Joachimiak at UTSW! Our new paper on the structure and function of an oligomer chaperone prohibiting polyglutamine aggregation has now been published in the journal Nature Communications. In it, we deploy a combination of solid-state NMR (ssNMR), solution NMR, cross-linking mass spectrometry (XL-MS) and other methods to visualise and understand these Hsp40 DnaJB8. This so-called co-chaperone teams up with Hsp70 to prevent polyglutamine aggregation in cells, but has been hard to understand. This is because it has itself a strong propensity to self-aggregate (or phase separate), even in a cellular context. Here we deploy a range of methods to identify and characterise an interesting and important feature of this multi domain protein. We show that the protein blocks its own activity when it is just “sitting around”, but can be activated due to environmental triggers. We figure out the specific amino acids are involved, and find that similar interactions are apparently present in other analogous chaperones. The intriguing possibility now arises that this self-blocking allows the chaperones to sit around in a “dormant” state until their action is (urgently?) needed. Then, perhaps, substrate proteins that could cause disease could be bound and that this binding interaction would quickly activate a bunch of these proteins. Further experiments will be needed to test this idea, and whether it can be used to control or activate these innate protective mechanisms as part of disease treatments in the future. Note that polyglutamine proteins aggregate in many incurable diseases, including Huntington’s disease (HD).

For more information, please read our new paper at the journal:
Ryder, B.D.; Matlahov, I.; Bali, S.; Vaquer-Alicea, J.; Van der Wel, P.C.A. & Joachimiak, L.A. (2021) Regulatory inter-domain interactions influence Hsp70 recruitment to the DnaJB8 chaperone. Nat. Commun. 12: 946 [DOI: https://doi.org/10.1038/s41467-021-21147-x]
Our work for this paper was made supported by the American NIH funding our polyglutamine research and instrumentation (grants GM112678 & OD012213-01). Nowadays, our polyglutamine research continues in Groningen with funding from CampagneTeam Huntington and CHDI. Note that this paper is published “open access” like all of our (recent) HD related work.
Note added:
The paper also received some online attention in some science-oriented news sites, based on press releases by the universities involved. This includes a highlight on the HD News website on April 20th.
New (e)book on membrane studies by solid-state NMR released online.
Now available online: we have contributed a chapter to a new (e)book on the topic of solid-state NMR studies of membranes and membrane proteins, edited by Frances Separovic and Marc-Antoine Sani (Univ. Melbourne, Australia). The edited volume “Solid state NMR. Applications in biomembrane structure.” was released in the IOP series in association with the Biophysical Society.
Our chapter (Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions [1]) looks at several studies that use ssNMR to probe peripheral membrane proteins. A key focus is on our own work on the mitochondrial protein cytochrome c, and how it binds to cardiolipin lipids during apoptosis, funded by the NIH/NIGMS [2]. In the chapter we try to summarise some of the practical challenges involved, along with potential solutions reported by ourselves and a few other research groups that studied other peripheral membrane proteins by ssNMR.
Cited references:
[1] Van der Wel, P.C.A. (2020) Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions. Chapter 10 in Solid-state NMR; applications in biomembrane structure. Edited by F. Separovic & M.-A. Sani; IOP Press (DOI 10.1088/978-0-7503-2532-5ch10)
[2] Li, M.; Mandal, A.; Tyurin, V. A.; DeLucia, M.; Ahn, J.; Kagan, V. E.; van der Wel, P. C. A. (2019) Surface-Binding to Cardiolipin Nanodomains Triggers Cytochrome c Pro-Apoptotic Peroxidase Activity via Localized Dynamics. Structure 2019, 27 (5), 806-815.e4. (DOI 10.1016/j.str.2019.02.007)
New publication on structure of mutant huntingtin protein from Huntington’s Disease
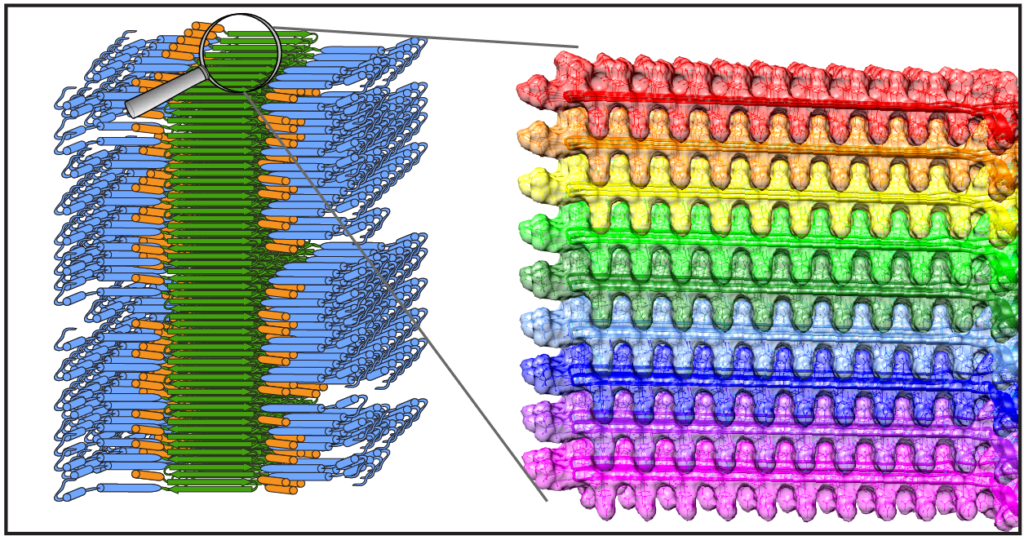
Congratulations to Dr. Jennifer Boatz and other team members for the acceptance and publication of a nice new paper on the structure of the misfolded mutant protein from Huntington’s disease. The paper is online at the Journal of Molecular Biology. Based on an integration of multiple techniques (NMR, EM, and X-ray diffraction), Jennifer assembled a new structural model of the protofilaments that make up the hierarchical fiber architecture of mutant huntingtin exon 1. This is a timely and important step forward in our understanding of the (mis)behaviour of the HD protein, and in particular how it forms pathogenic inclusions and protein aggregates.
In this new paper and prior work we (and others) have seen that the mutant protein is prone to form a collection of different kinds of aggregates, with each their own structure and functional properties. This is of substantial interest from a disease perspective, as these “functional properties” can encompass different degrees of neurotoxic properties. Surprisingly, Jennifer shows in this paper that one contributor to, or trigger of, huntingtin polymorphism is the concentration of the protein. Further studies will have to explore whether or how this finding impacts efforts to replicate cellular behaviour of the protein in vitro, and how it may affect the toxic properties of the aggregated protein states.
Reference:
Protofilament Structure and Supramolecular Polymorphism of Aggregated Mutant Huntingtin Exon 1. Boatz, J.C., Piretra, T., Lasorsa, A., Matlahov, I., Conway, J.F. & Van der Wel, P.C.A. (2020) J. Mol. Biol., 432(16): 4722-4744. [DOI] (Open Access)
Funding & support: The underlying research in this paper was enabled by funding support from the American NIH/NIGMS (grant R01 GM112678) and funding from the CampagneTeam Huntington in the Netherlands. For more information on the disease, see also our HD page and the CTH website. Other support came from the University of Groningen and the University of Pittsburgh.
Note added: Our Institute also highlighted this paper in a nice summary posted on the Zernike Institute website.
New review paper on ssNMR studies of polysaccharide hydrogels
PhD student Mustapha El Hariri El Nokab has put together a nice new review of solid-state NMR studies of polysaccharide hydrogels, which is now out in the journal Carbohydrate Polymers. This review is part of his research project that make use of solid-state NMR (and other tools) to look at functional polysaccharide hydrogels. This new research direction in the lab also constitutes part of our participation in the new Physics of Cancer (www.phycan.nl) initiative of the Zernike Institute for Advanced Materials.
Please find the open-access published paper at the journal Carbohydrate Polymers:
New Publication on Lipid Oxidation
Now online in the journal Free Radical Biology and Medicine: a new review and perspective article by our collaborator Valerian Kagan (Univ. Pittsburgh). The paper (titled “Redox phospholipidomics of enzymatically generated oxygenated phospholipids as specific signals of programmed cell death”) examines the role of controlled lipid oxidation as a source of vital cellular signals. In particular it reviews recent work showing how cardiolipins and phosphatidylethanolamine lipids are oxidised by enzymes, as triggers of apoptosis and ferroptosis. Interestingly, the enzymatically generated oxidised species are distinct from those generated by spontaneous peroxidation, which may be important for the regulatory role of these species.
For more details, read the whole article here:
Kagan et al. (2020) “Redox phospholipidomics of enzymatically generated oxygenated phospholipids as specific signals of programmed cell death” Free Radical Biology and Medicine, Vol. 147, pp. 231-241
Publication: New review article on the structural biology of Huntington’s disease.
Now online at the journal Experimental Biology and Medicine: our new review article summarizing recent contributions from solid-state NMR and electron microscopy to further our understanding of the (mis)behavior of the mutant proteins behind Huntington’s disease.
Citation:
I. Matlahov & P.C.A. van der Wel (2019) Conformational studies of pathogenic expanded polyglutamine protein deposits from Huntington’s disease. Exp. Biol. Med. in press; DOI: 10.1177/1535370219856620
Publication: cytochrome c-cardiolipin complexes as pro-apoptotic lipid peroxidase.
Our first paper of 2019 has just appeared online in the journal Structure. It describes the very nice solid-state NMR studies performed by Dr. Mingyue Li, on a protein-lipid complex involved in the early stages of mitochondrial apoptosis. Together with our collaborators in the groups of Valerian Kagan and Jinwoo Ahn (University of Pittsburgh), she looked at the structure and function of the peroxidase active cytochrome c in its membrane-bound state. For more details on the findings, including how the lipid substrate cardiolipin forms membrane nano domains and acts as a dynamic regulator, please see the paper at the journal. This work was made possible by NIH funding (NIGMS R01 GM113908).
Reference:
Surface-Binding to Cardiolipin Nanodomains Triggers Cytochrome c Pro-apoptotic Peroxidase Activity via Localized Dynamics. Li, M., Mandal, A., Tyurin, V.A., Delucia, M., Ahn, J., Kagan, V.E., & Van der Wel, P.C.A. (2019) Structure, in press [URL]
Additional information/press info:
Publication: new collaborative paper on DNP/ssNMR-enabled studies at natural abundance (JACS).
Congrats to Talia, Jennifer and Irina, as well as our international network of collaborators on the acceptance of an exciting new paper in JACS. It describes the power of dynamic nuclear polarization (DNP) to enable multidimensional solid-state NMR studies of polyglutamine-expanded huntingtin exon 1 fibrils. Importantly in these experiments we did not apply stable-isotope labeling to these protein deposits that are implicated in Huntington’s disease. This approach enables great structural data and can hopefully prove applicable to many more (types of) samples.
More info to follow, but for now: Read the accepted paper here at the journal.
Title: Structural fingerprinting of protein aggregates by DNP-enhanced solid-state NMR at natural isotopic abundance
Authors: Adam N. Smith, Katharina Märker, Talia Piretra, Jennifer C. Boatz, Irina Matlahov, Ravindra B. Kodali, Sabine Hediger, Patrick C.A. van der Wel, and Gaël De Paëpe
DOI: 10.1021/jacs.8b09002
Publication: Dynamics-based spectral editing – DYSE SSNMR.
Our new review article discussing the concept of dynamics-based spectral editing (DYSE) in ssNMR is now available online in the journal Methods. In it, we describe how the dynamics-sensitive pulse sequences enable the filtering out (or spectral editing) of signals based on their differences in dynamics. Applications by ourselves and many other groups are discussed. Notably, these applications range across a huge swatch of different sample types, going from designer peptide nano materials to whole tissues and even living organisms.
Citation:
- Hidden motions and motion-induced invisibility: dynamics-based spectral editing in solid-state NMR. Matlahov, I., Van der Wel, P.C.A. Methods, in press [URL]
Publication: New applications of solid-state NMR in structural biology. (review)
Our new paper discussing the role of modern solid-state NMR in integrated structural biology is now published in Emerging Topics in Life Sciences. It provides a review of a selection of recent solid-state NMR studies from (mostly) the last decade. Not only does it give a review of notable 3D structures obtained by solid-state NMR, but it also examines contributions going beyond static protein structures. This includes the characterization of biologically relevant dynamics, and the pinpointing of crucial intermolecular interactions.
Reference:
- Patrick C.A. van der Wel (2018) “New applications of solid-state NMR in structural biology.” Emerging Topics in Life Sciences, Vol. 2, pp. 57–67
Access:
- DOI: 10.1042/ETLS20170088
- Request reprints by email: vanderwel@pitt.edu.
- Open Access PDF for download: OA PDF
