Tag: polyQ
Publication: New paper about photochemical approaches to studying polyQ protein aggregation.
Congratulations to PhD student Raffaella Parlato, Dr. Jana Volaric, and their collaborators, on the publication of a nice new paper in the Journal of the American Chemical Society. This is the final result of an idea from some years ago, which came together very nicely thanks to a great team of collaborators. The goal of this work was to explore the idea of putting polyglutamine aggregation under some degree of photo-control. In prior work, it has been shown that b-hairpin formation is a key step in the aggregation process of expanded polyQ proteins. Azobenzene-based groups can be used to favor or disfavor beta turn structures under the influence of light, but prior ways of implementing this have been limited in various ways. A new amino acid analogue is introduced with more favorable structural and photochemical properties. And, we tested it in the context of polyQ peptides, seeing it indeed modulating the conformation of resulting aggregates (seen by ssNMR on unlabeled peptide fibrils!). For more details, see the paper. It is open access, so easily accessible.
Citation:
Raffaella Parlato, Jana Volarić, Alessia Lasorsa, Mahdi Bagherpoor Helabad, Piermichele Kobauri, Greeshma Jain, Markus S. Miettinen, Ben L. Feringa*, Wiktor Szymanski*, and Patrick C. A. van der Wel* (2024) J. Am. Chem. Soc. 2024, 146, 3, 2062–2071. Photocontrol of the β-Hairpin Polypeptide Structure through an Optimized Azobenzene-Based Amino Acid Analogue. DOI: https://doi.org/10.1021/jacs.3c11155
Publication: Review article on polyglutamine protein analysis by solid-state NMR spectroscopy.
Our new review article summarizing progress in polyglutamine protein studies by solid-state NMR is now available on the website of the journal Biochemical Society Transactions. This is an invited mini-review that was requested to cover the progress made in our understanding the aggregation and misfolding of mutant polyglutamine proteins implicated in diseases such as Huntington’s disease and several versions of spinocerebellar ataxia (SCA). We describe how studies from ssNMR has been used to better understand the structure of these protein aggregates, and thus helps us analyze how the aggregation process occurs and how aggregates may play a role in these diseases. The paper is published open-access, so you should be able to read it to see all the details. (Naturally, the format of the mini-review limited the word count and meant that much had to be omitted)
Citation:
Patrick C.A. van der Wel; Solid-state nuclear magnetic resonance in the structural study of polyglutamine aggregation. Biochem Soc Trans 2024; BST20230731. doi: https://doi.org/10.1042/BST20230731
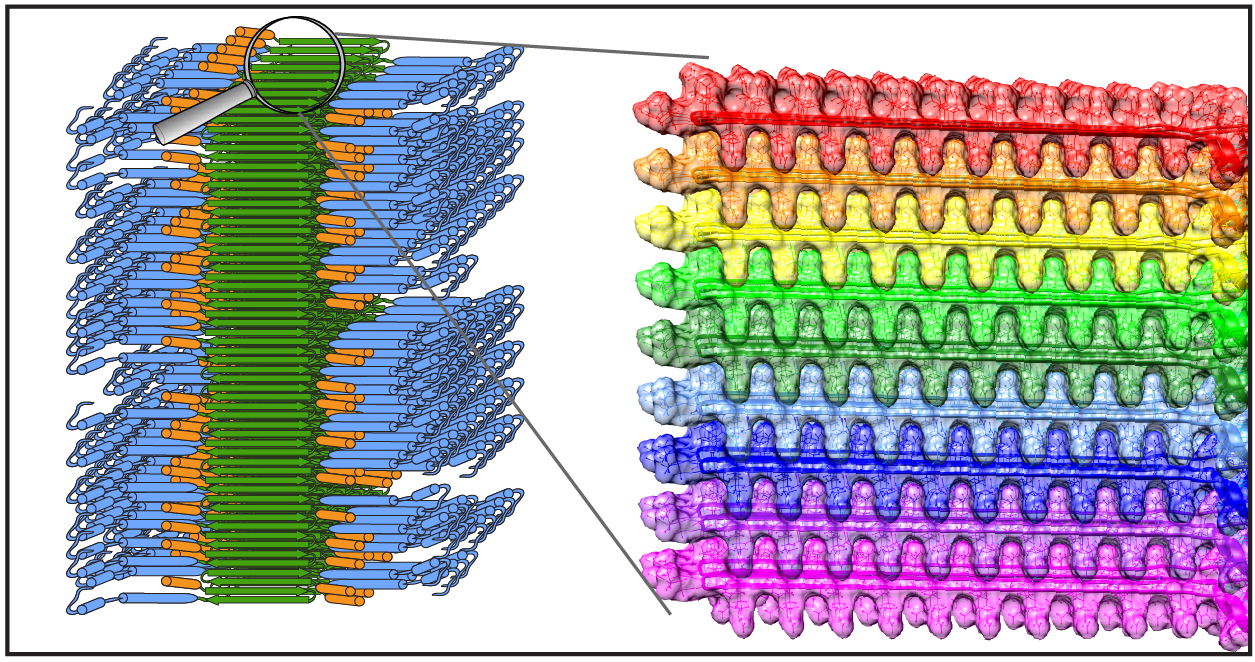
Publication: Dynamics-based spectral editing to see (fiber) surfaces by solid-state NMR.
Congratulations to Dr. Irina Matlahov and (alum) Dr. Jennifer Boatz on the publication of their new paper in the Journal of Structural Biology X. The paper is entitled “Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils“. It describes our latest research on the misfolded protein deposits associated with Huntington’s disease (HD), specifically looking at what happens on the surface of these protein fibrils.
In previous work we have studied the structure of these nanometer-sized fibrils formed by mutant huntingtin’s exon 1 fragment, using ssNMR, EM and other methods. In our earlier studies we used dynamics-sensitive NMR techniques to look at the core of these fibrils, taking advantage of its very rigid structure. Conversely, we had looked at super flexible protein parts that are away from the rigid core. Those types of techniques have been a popular tool for seeing rigid or flexible parts of (aggregated) proteins. However, in the current paper we try to look for protein parts with intermediate mobility. Of particular interest are those residues that form the surface of the rigid fibril core: residues that are somewhat immobilized by proximity to the core proper, but are somewhat mobilized by their interactions with water solvent surrounding the fibril. This part of the protein fibrils can be especially interesting, as it is the part seen by protein-targeting antibodies, PET ligands and protein-protein interaction partners in affected neurons.
The paper discusses how we can probe the impact of the water interactions with the surface through variable temperature ssNMR. However, it also shows the limitations of this traditional approach. Instead we advocate for a new type of dynamic filtering experiment that selectively shows the signals of semi-rigid/semi-mobile residues (such as those on the fiber surface). This technique (IMS-DYSE) is complementary to traditional DYSE methods that select rigid or flexible sites. For more details, please see the paper (linked below). We expect to combine this dynamic filtering technique with other types of pulse sequences, to probe in more detail the features of the fiber surface.
Note that this technique proved especially useful for our work on polyglutamine protein fibrils, as in this protein system we have a core made up of glutamine residues, providing an overwhelming strong NMR signal that masks the resonances of the surface glutamines. So, to see the surface, it is essential to find a way to suppress the core signals, leaving only those of the surface residues. The paper shows that this indeed works, and we can for the first time use this technique to distinguish the core and surface glutamines. With this technique in hand, we can foresee further studies of surface-specific features, such as the binding sites for targeted antibodies and/or amyloid-binding dyes.
The paper is available online at the journal, and has the following citation:
I. Matlahov, J.C. Boatz, P.C.A. van der Wel (2022) Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils. J. Struct. Biol. X, vol. 6, 100077. DOI: 10.1016/j.yjsbx.2022.100077
PS. Some of the techniques and results from this publication were also discussed during a prior online seminar in the MIT ssNMR/DNP zoominar series. You can view that video here.
News: Dutch-language radio
Recently our research was covered (briefly) on a Dutch radio station, featuring an interview with Patrick. It is mostly about our work applying ssNMR to studying how proteins aggregate in Huntington’s disease. You can find the recording online at the NPO radio website. More information about our Huntington disease research can also be found on this page, and in two recent webinars.

Upcoming event: BPS networking event on polyQ biophysics
Hereby an early announcement of an upcoming online event hosted by our group and the group of Markus Miettinen: “Biophysics of polyglutamine aggregation: how does it start and how does it end?“
This is an online event is organized in context of the networking event series sponsored and organized by the Biophysical Society (BPS). Our event aims to bring together international scientists working on the biophysics of polyglutamine (polyQ) protein aggregation. The event will take place on July 6th 2022, from 15:00-18:00 CET.
More information can be found at the page from the BPS or our own institute. An online registration page is planned to online at the BPS, see below. If you want more information in the mean time, please contact us by email.
Update May 2022: the registration page is now active at the BPS. The link is here. Note that this does require a free BPS account to work. Let us know if you have any issues or questions.
Webinar: Structural biology of Huntington’s disease.
On May 15th 2021, Patrick gave an invited lecture in the online webinar series on the Molecular Bases of Proteinopathies hosted by the Ramamoorthy lab at the University of Michigan. Patrick talked about our research on polyglutamine protein aggregation and the structural biology of Huntington’s disease.The seminar title was “The Structural Biology of Protein Misfolding in Huntington’s disease”. If you are interested, the seminar has been posted to YouTube, where it can be viewed here.