Blog Posts
Publication: Review article on polyglutamine protein analysis by solid-state NMR spectroscopy.
Our new review article summarizing progress in polyglutamine protein studies by solid-state NMR is now available on the website of the journal Biochemical Society Transactions. This is an invited mini-review that was requested to cover the progress made in our understanding the aggregation and misfolding of mutant polyglutamine proteins implicated in diseases such as Huntington’s disease and several versions of spinocerebellar ataxia (SCA). We describe how studies from ssNMR has been used to better understand the structure of these protein aggregates, and thus helps us analyze how the aggregation process occurs and how aggregates may play a role in these diseases. The paper is published open-access, so you should be able to read it to see all the details. (Naturally, the format of the mini-review limited the word count and meant that much had to be omitted)
Citation:
Patrick C.A. van der Wel; Solid-state nuclear magnetic resonance in the structural study of polyglutamine aggregation. Biochem Soc Trans 2024; BST20230731. doi: https://doi.org/10.1042/BST20230731
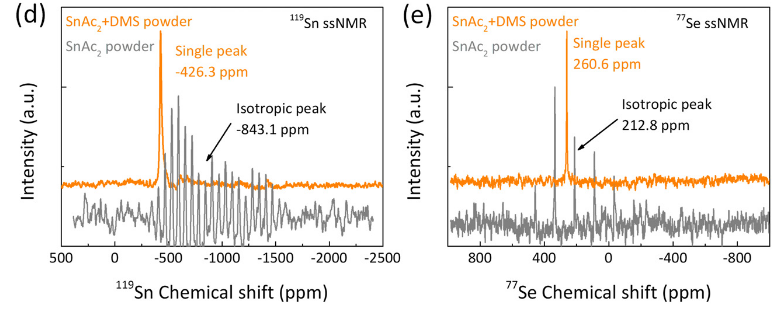
Publication: collaborative paper on perovskite-related materials, with Sn and Se ssNMR
Congrats to our collaborators from the group of Maria Loi and others for the publication of their new paper on in situ SnSe deposition as passivation for scalable and stable quasi-2D lead–tin perovskite solar cells, in the journal Energy & Environmental Science. Dr. Lasorsa in our group contributed tin and selenium ssNMR analysis to this interdisciplinary paper. For more information see the paper at the journal.
Publication:
Chen L, Tekelenburg EK, Gahlot K, Pitaro M, Xi J, Lasorsa A, et al. In situ SnSe deposition as passivation for scalable and stable quasi-2D lead–tin perovskite solar cells. Energy Environ Sci. 2023;10.1039.D3EE02507A.
Educational webinar on the use of ssNMR to measure dihedral angles.
Recently, Patrick gave an online lecture in the online webinar tutorial series of the Global NMR Discussion Meetings series (episode 70!). In this online lecture he introduced and discussed approaches to measure torsion angles (or dihedral angles) using advanced solid-state NMR spectroscopy. He discussed the basic idea of how these are implemented in ssNMR, but also how such structural data can be a useful complement to more traditional inter-atomic distance information obtained by ssNMR. You can now watch the recording of this lecture on Youtube, via this link.
Want to learn even more?
This lecture is connected to our recent review article on the same topic, which can be found online at: Frontiers | Dihedral Angle Measurements for Structure Determination by Biomolecular Solid-State NMR Spectroscopy (frontiersin.org)
Publication: Dynamics-based spectral editing to see (fiber) surfaces by solid-state NMR.
Congratulations to Dr. Irina Matlahov and (alum) Dr. Jennifer Boatz on the publication of their new paper in the Journal of Structural Biology X. The paper is entitled “Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils“. It describes our latest research on the misfolded protein deposits associated with Huntington’s disease (HD), specifically looking at what happens on the surface of these protein fibrils.
In previous work we have studied the structure of these nanometer-sized fibrils formed by mutant huntingtin’s exon 1 fragment, using ssNMR, EM and other methods. In our earlier studies we used dynamics-sensitive NMR techniques to look at the core of these fibrils, taking advantage of its very rigid structure. Conversely, we had looked at super flexible protein parts that are away from the rigid core. Those types of techniques have been a popular tool for seeing rigid or flexible parts of (aggregated) proteins. However, in the current paper we try to look for protein parts with intermediate mobility. Of particular interest are those residues that form the surface of the rigid fibril core: residues that are somewhat immobilized by proximity to the core proper, but are somewhat mobilized by their interactions with water solvent surrounding the fibril. This part of the protein fibrils can be especially interesting, as it is the part seen by protein-targeting antibodies, PET ligands and protein-protein interaction partners in affected neurons.
The paper discusses how we can probe the impact of the water interactions with the surface through variable temperature ssNMR. However, it also shows the limitations of this traditional approach. Instead we advocate for a new type of dynamic filtering experiment that selectively shows the signals of semi-rigid/semi-mobile residues (such as those on the fiber surface). This technique (IMS-DYSE) is complementary to traditional DYSE methods that select rigid or flexible sites. For more details, please see the paper (linked below). We expect to combine this dynamic filtering technique with other types of pulse sequences, to probe in more detail the features of the fiber surface.
Note that this technique proved especially useful for our work on polyglutamine protein fibrils, as in this protein system we have a core made up of glutamine residues, providing an overwhelming strong NMR signal that masks the resonances of the surface glutamines. So, to see the surface, it is essential to find a way to suppress the core signals, leaving only those of the surface residues. The paper shows that this indeed works, and we can for the first time use this technique to distinguish the core and surface glutamines. With this technique in hand, we can foresee further studies of surface-specific features, such as the binding sites for targeted antibodies and/or amyloid-binding dyes.
The paper is available online at the journal, and has the following citation:
I. Matlahov, J.C. Boatz, P.C.A. van der Wel (2022) Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils. J. Struct. Biol. X, vol. 6, 100077. DOI: 10.1016/j.yjsbx.2022.100077
PS. Some of the techniques and results from this publication were also discussed during a prior online seminar in the MIT ssNMR/DNP zoominar series. You can view that video here.
News: Dutch-language radio
Recently our research was covered (briefly) on a Dutch radio station, featuring an interview with Patrick. It is mostly about our work applying ssNMR to studying how proteins aggregate in Huntington’s disease. You can find the recording online at the NPO radio website. More information about our Huntington disease research can also be found on this page, and in two recent webinars.

Publication: SSNMR of alginate hydrogel (re)hydration
A new open-access publication by Mustapha and collaborators has been published in the journal Food Hydrocolloids, describing how he used various ssNMR measurements to probe alginate hydrogel structure and (re)hydration. Alginates can be cross-linked with calcium to form hydrogels, which are used in many different types of applications. This includes their use in slow-release drug delivery as well as food/nutritional applications. In this new paper, Mustapha shows how 1H, 2H and 13C ssNMR measurements can be used to see how these hydrogels are structured, but especially also how they are hydrated by the aqueous solvent. A key feature of the work is the use of 1H MAS NMR to detect the water molecules in the hydrogel macropores as well as those waters directly interacting with the alginate. The two types of water molecules form distinct ‘pools’ in the hydrogel, which can be nicely distinguished by their different molecular motion. The latter is detected via different relaxation measurements, in these NMR experiments.
To read more:
El Hariri El Nokab, M..; Lasorsa, A.; Sebakhy, K. O.; Picchioni, F.; van der Wel, P. C. A. Solid-State NMR Spectroscopy Insights for Resolving Different Water Pools in Alginate Hydrogels. Food Hydrocolloids 2022, 107500.
Publication: New paper on mitochondrial protein-lipid interactions published in PNAS.
Congratulations to lab alumns Dr. Abshishek Mandal and Dr. Jennifer Boatz, as well as our collaborators in the USA and Spain! A new collaborative paper on mitochondrial protein-lipid interactions has just been published in the journal PNAS. In this multidisciplinary work we studied how the protein Drp1 binds the special mitochondrial lipid cardiolipin in order to do its job managing the proper fission of mitochondrial membrane. Our collaborator Rajesh Ramachandran (at Case Western) coordinated a wide array of experimental and computational approaches to determine how Drp1’s “variable domain” (VD) binds cardiolipin. Along the way, two apparent CL-binding motifs were detected, which are seemingly shared by other CL-binding proteins. Their mutation disrupts CL binding and also the proper management of mitochondrial morphology, pointing to the importance of CL-based signals and interactions in this vital cellular process.
Experimentally, our group contributed various solid-state NMR measurements of the lipids, with which we probe the specificity of the interactions and also observe how the protein modulates the lipid bilayer itself. Moreover, we were involved in the sequence/structure analysis: sequence analysis of the “disordered” VD domain pinpointed those parts of the structure most likely to engage in lipid-driven folding upon membrane binding. The “MoRF” motifs (“molecular recognition features”) indeed appear to be involved in CL binding, based on NMR and mutational studies. In Drp1 they are intimately involved in a folding transition of the VD domain upon membrane binding. It will be interesting to see how widespread these CL-binding motifs (CBMs) are in other CL-binding proteins.
Reference:
[1] Mahajan M, Bharambe N, Shang Y, Lu B, Mandal A, Madan Mohan P, et al. NMR identification of a conserved Drp1 cardiolipin-binding motif essential for stress-induced mitochondrial fission. Proc Natl Acad Sci USA2021;118:e2023079118. https://doi.org/10.1073/pnas.2023079118.
New (e)book on membrane studies by solid-state NMR released online.
Now available online: we have contributed a chapter to a new (e)book on the topic of solid-state NMR studies of membranes and membrane proteins, edited by Frances Separovic and Marc-Antoine Sani (Univ. Melbourne, Australia). The edited volume “Solid state NMR. Applications in biomembrane structure.” was released in the IOP series in association with the Biophysical Society.
Our chapter (Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions [1]) looks at several studies that use ssNMR to probe peripheral membrane proteins. A key focus is on our own work on the mitochondrial protein cytochrome c, and how it binds to cardiolipin lipids during apoptosis, funded by the NIH/NIGMS [2]. In the chapter we try to summarise some of the practical challenges involved, along with potential solutions reported by ourselves and a few other research groups that studied other peripheral membrane proteins by ssNMR.
Cited references:
[1] Van der Wel, P.C.A. (2020) Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions. Chapter 10 in Solid-state NMR; applications in biomembrane structure. Edited by F. Separovic & M.-A. Sani; IOP Press (DOI 10.1088/978-0-7503-2532-5ch10)
[2] Li, M.; Mandal, A.; Tyurin, V. A.; DeLucia, M.; Ahn, J.; Kagan, V. E.; van der Wel, P. C. A. (2019) Surface-Binding to Cardiolipin Nanodomains Triggers Cytochrome c Pro-Apoptotic Peroxidase Activity via Localized Dynamics. Structure 2019, 27 (5), 806-815.e4. (DOI 10.1016/j.str.2019.02.007)
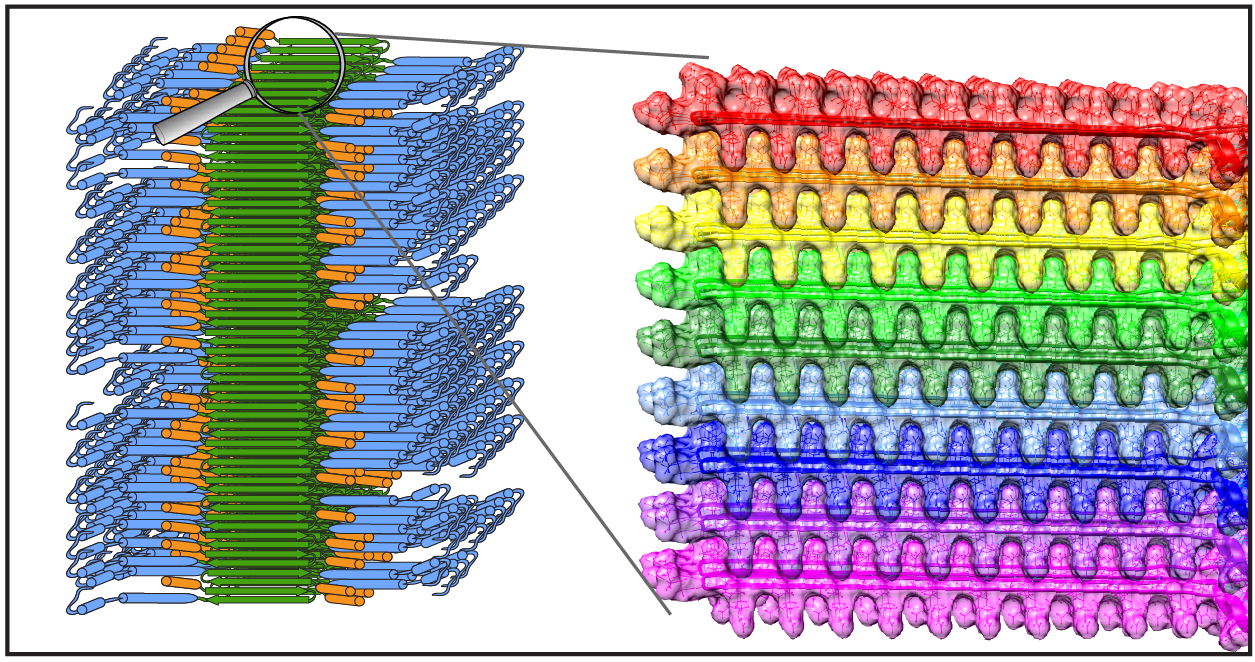
New publication on structure of mutant huntingtin protein from Huntington’s Disease
Congratulations to Dr. Jennifer Boatz and other team members for the acceptance and publication of a nice new paper on the structure of the misfolded mutant protein from Huntington’s disease. The paper is online at the Journal of Molecular Biology. Based on an integration of multiple techniques (NMR, EM, and X-ray diffraction), Jennifer assembled a new structural model of the protofilaments that make up the hierarchical fiber architecture of mutant huntingtin exon 1. This is a timely and important step forward in our understanding of the (mis)behaviour of the HD protein, and in particular how it forms pathogenic inclusions and protein aggregates.
In this new paper and prior work we (and others) have seen that the mutant protein is prone to form a collection of different kinds of aggregates, with each their own structure and functional properties. This is of substantial interest from a disease perspective, as these “functional properties” can encompass different degrees of neurotoxic properties. Surprisingly, Jennifer shows in this paper that one contributor to, or trigger of, huntingtin polymorphism is the concentration of the protein. Further studies will have to explore whether or how this finding impacts efforts to replicate cellular behaviour of the protein in vitro, and how it may affect the toxic properties of the aggregated protein states.
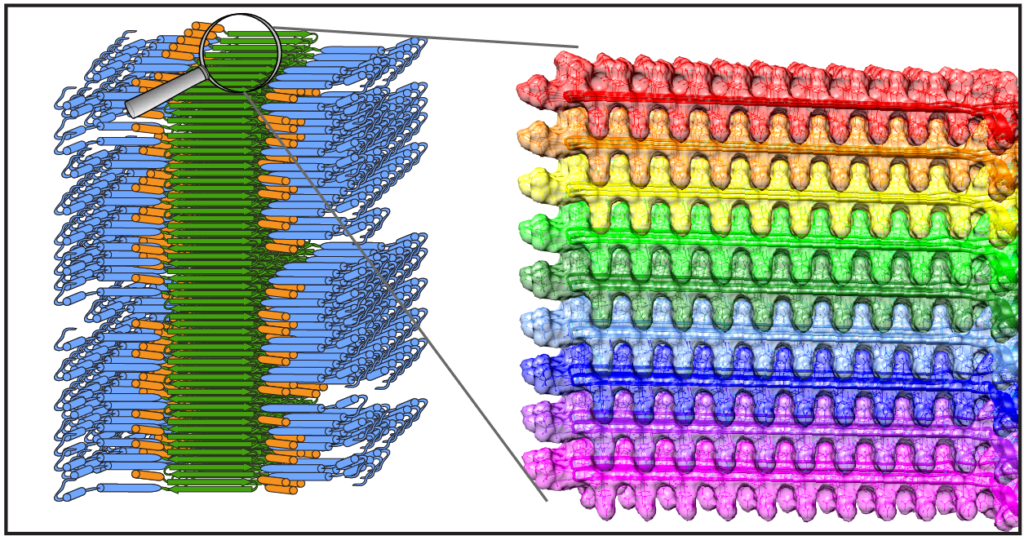
Reference:
Protofilament Structure and Supramolecular Polymorphism of Aggregated Mutant Huntingtin Exon 1. Boatz, J.C., Piretra, T., Lasorsa, A., Matlahov, I., Conway, J.F. & Van der Wel, P.C.A. (2020) J. Mol. Biol., 432(16): 4722-4744. [DOI] (Open Access)
Funding & support: The underlying research in this paper was enabled by funding support from the American NIH/NIGMS (grant R01 GM112678) and funding from the CampagneTeam Huntington in the Netherlands. For more information on the disease, see also our HD page and the CTH website. Other support came from the University of Groningen and the University of Pittsburgh.
Note added: Our Institute also highlighted this paper in a nice summary posted on the Zernike Institute website.
Competitive PhD Scholarships call open (deadline April 1st)
Our Institute and Faculty have opened up a competitive call for PhD scholarship applications. Our solid-state NMR group participates in one of the research theme areas, designated as “Advanced Materials”, which spans topics from physics of life, via bio-inspired materials, to materials and much more. The process is described in some detail on the RuG website. Briefly, applicants (with a MSc degree) are expected to contact a PI/supervisor (immediately!) and develop a fitting project to submit. The submission deadline of an initial idea (few hundred words) is due by April 1st 2020!
Suitable ideas are expected to fit within designated topics areas, described on the website here and here. Note especially also the 2nd link, as it contains important detailed information.T
Interested in this? Please contact us as soon as possible, to meet the tight deadlines. Together we can consider topics that range from self-assembling bio-inspired materials, Physics of Cancer, non-biological materials and other topics. Projects will be highly interdisciplinary as the involvement of a second supervisor is also required, with a distinct and complementary expertise.
Publication: Impact of aggregation inhibitors on protein aggregation and aggregate toxicity.
Congratulations to Greeshma Jain, Marina Trombetta-Lima and the whole team of co-authors for the publication of our most recent paper on Huntington’s disease (HD), in the journal Nature Communications. This paper is actually the fourteenth peer-reviewed publication from the lab on HD research, since our first paper in 2011. It was enabled by crucial funding from the CampagneTeam Huntington foundation, who supported our HD research program when the Van der Wel group moved back to the Netherlands.
In this new report we describe our use of structural analysis (NMR and EM) to look how aggregation-inhibiting small molecules (polyphenols) change the structure of protein aggregates formed by fragments of the huntingtin protein. We observed that these molecules are effective at slowing down huntingtin exon 1 aggregation, even at sub-stoichiometric concentrations. However, aggregates still formed. Notably, these ‘break-through’ aggregates seem to adopt a different conformation, resulting from a change in the aggregation mechanism. Moreover, we saw that the apparent toxic or pathogenic effects of these aggregates differ from normal aggregates, when we exposed neuronal cells to them. What precisely makes the differ in the reduced toxicity remains a bit unclear, requiring further research.
However, a notable and more general finding is that we need to pay attention to changes in aggregate polymorphs when developing aggregation inhibitors. Here we see a reduced toxic effect, but it seems conceivable that certain inhibitors may redirect the aggregation pathway to more toxic species. Further research would be important for better understanding the interplay of delayed aggregation, changes in aggregate morphology and cellular impacts of protein aggregates.
For more info, please see the actual paper at the journal, with citation below:
Jain G, Trombetta-Lima M, Matlahov I, Ribas HT, Chen T, Parlato R, Portale G, Dolga AM, Van der Wel PCA. Inhibitor-based modulation of huntingtin aggregation mechanisms mitigates fibril-induced cellular stress. Nat Commun. 2025 Apr 15;16(1):3588.
Webinar: GlycoScience Webinar on polysaccharide ssNMR research
On 27 March Patrick gave an online presentation in the EGC webinar series, on the topic of using solid-state NMR (and isotope labeling) to study polysaccharides in various contexts. The title of the presentation was: “Labeling and solid-state NMR studies of hyaluronic acid and other polysaccharides: hydrogels, nanoparticles and more”
EGC Webinar #86 Glycochemistry (March 27, 4 pm CET)
Patrick van der Wel (Zernike Institute for Advanced Materials, University of Groningen, the Netherlands)
“Labeling and solid-state NMR studies of hyaluronic acid and other polysaccharides: hydrogels, nanoparticles and more”
A recording of the lecture is visible via YouTube. In the presentation he referred to several published stories, that you can access via our publications list page.

Publication: Peptide-based self-assemblers and self-replicators (with insights from ssNMR).
Congratulations to our collaborators in the group of Sijbren Otto (RUG), and Alessia, for the new paper in the journal Chem. This report discusses how self-assembling and self-replicating molecules can borrow principles from polypeptide amyloid formation. In particular, the concept of ‘steric zippers’ proves to be highly relevant – an idea from the amyloid field that comes out of seminal work by the group of David Eisenberg (UCLA). Our contributions in this new paper relate to the use of 1D and 2D solid-state NMR analysis of the self-assembled materials, in samples where targeted isotope-labeled amino acids were incorporated. With these NMR studies we were able to probe the structure and symmetry of the assembled state, giving some insights into the fibril architecture. See also the figure below, from the paper’s SI.
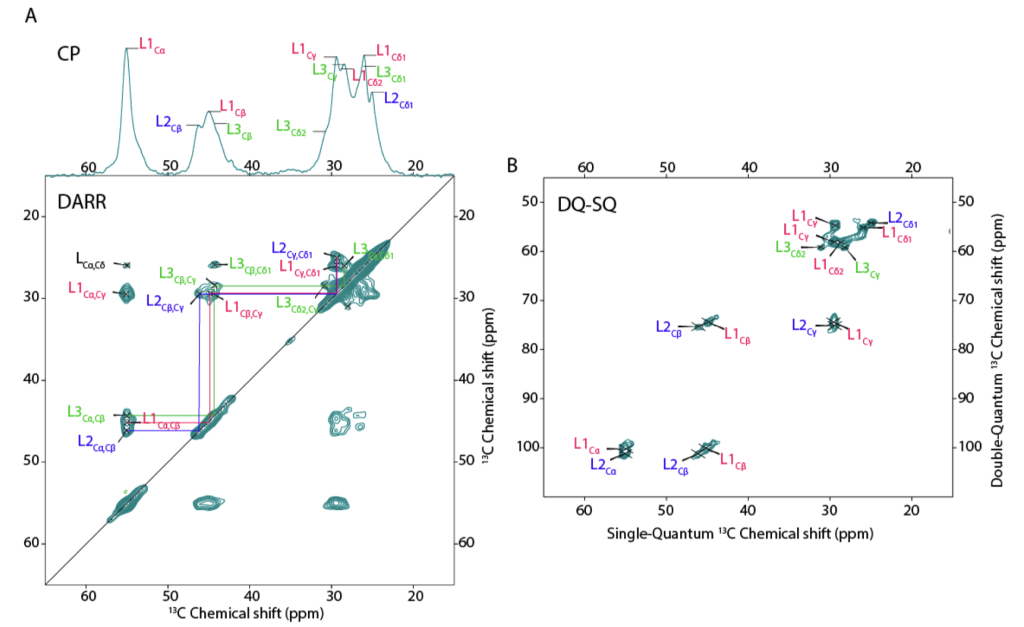
Citation: Eleveld, M.J., Wu, J., Liu, K., Ottelé, J., Markovitch, O., Kiani, A., et al. (2025) Departure from randomness: Evolution of self-replicators that can self-sort through steric zipper formation. Chem 102374 https://doi.org/10.1016/j.chempr.2024.11.012
PS. Conceptually and practically, this work has similarities to our earlier JACS paper with the group of Nat Rosi at the University of Pittsburgh, in which we helped detail the steric zipper motifs in a different class of peptide-chimera fibril-forming molecules. Also that paper may be interesting to revisit! [DOI https://doi.org/10.1021/jacs.6b07322]
Publication: Structure of protein aggregates implicated in Huntington disease
Congratulations to Irina, Greeshma, and Raffaella, together with our international network of collaborators, on a newly published paper that came out in Nature Communications in Dec 2024, in a special collection on the Biology of rare genetic disorders. In this collaborative work, we looked at the structure of proteins behind Huntington’s disease (HD), enabled by crucial funding from the CampagneTeam Huntington and the CHDI Foundation. HD is an inherited neurodegenerative disease, with patients having a mutated form of the huntingtin (HTT) gene. The mutation affects a repeating sequence of CAG codons that has been expanded in HD patients. This translates to huntingtin (HTT) proteins featuring an expanded polyglutamine (or polyQ) segment. Most people have about 20 residues in their HTT polyglutamine segment. Patients have more than 35. Postmortem analysis of HD patient brains reveals the presence of protein clumps (aggregates or inclusions) that contain short fragments of the mutated HTT protein.
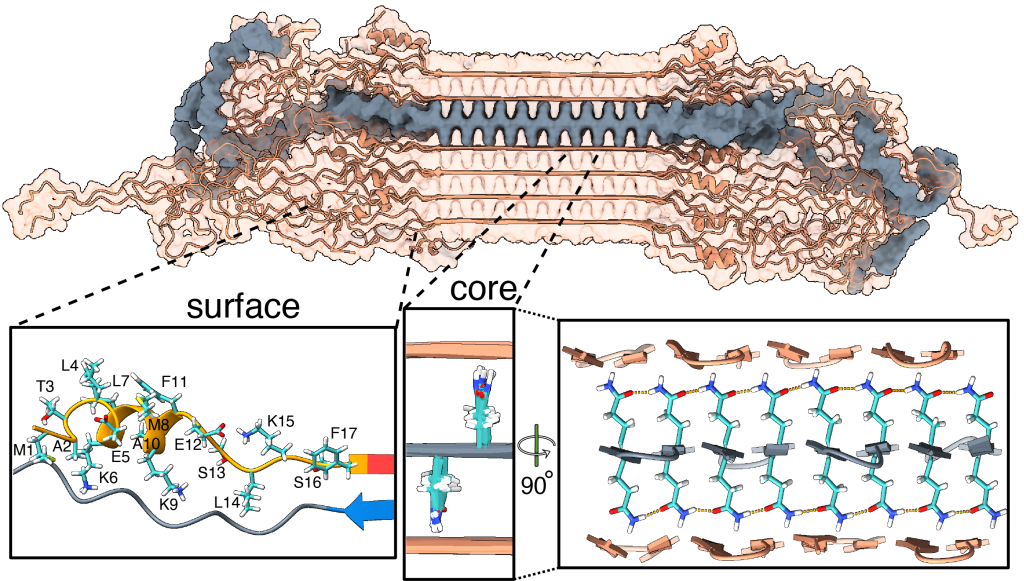
In past work we (and others) have studied how HTT fragments also form aggregates when you make the proteins in the lab, and do so more and more aggressively as you expand their polyglutamine segments. Although the precise role of these protein clumps in the disease remains under active debate, there is a consensus that they represent an important biomarker of the disease. Until now, we have lacked a solid understanding of the molecular conformation of the HTT protein in these clumps. For some years now my group (with various collaborators) has been trying to address this question.
Our new paper (together with the team of Markus Miettinen, University of Bergen) introduces the first atomic-level description of the HTT fragments that are increasingly thought to be relevant to the disease. We worked closely together to combine ssNMR data with results from other techniques to build a comprehensive and integrative model that visualizes these protein fibrils in atomic detail. Key aspects of the resulting structure are of course informed and determined by our experimental findings. However, the modeling also unveiled important and useful new insights.
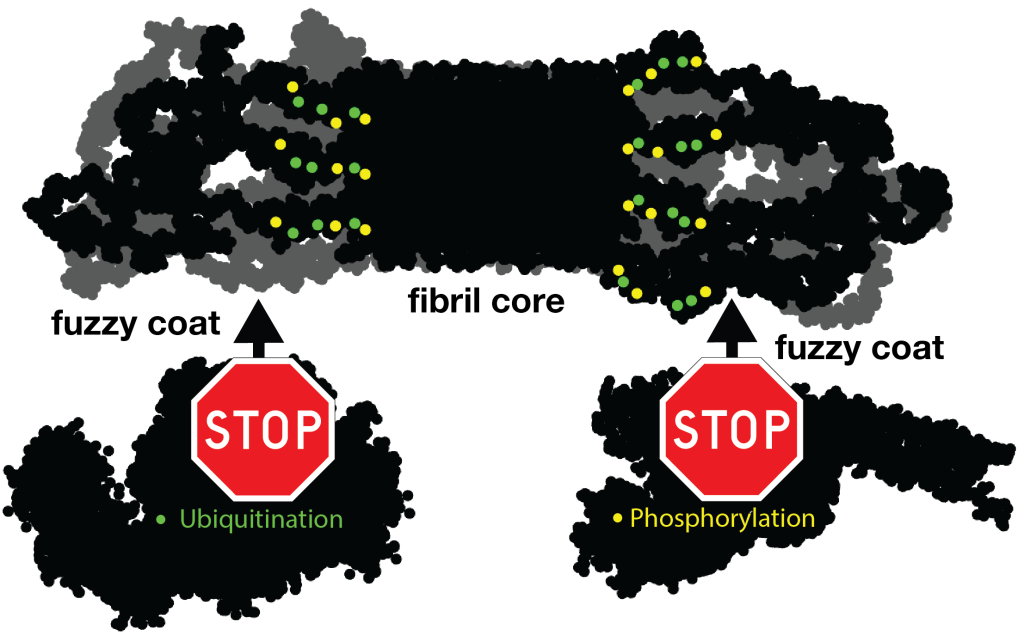
For instance, let’s look at the fuzzy coat that covers these HD amyloid fibrils. Regions outside the mutated polyQ stretch (known as ‘flanking segments’) form a disordered, but densely packed, brush on two sides of a roughly rectangular fibril core. We discuss in the paper how the dense packing reduces or prohibits interactions of other proteins with the flanking segments. This limits the ability of kinases to phosphorylate the aggregated proteins; and similarly for poly-ubiquitination. This may be seen as bad news, as both processes may be used to enable destabilization or destruction of the aggregates.
However, these brushes cover only two sides of the fibril core. The other two sides feature an extensive interface directly between the polyQ amyloid core and the surrounding aqueous phase. In laboratory experiments, it is this type of interface that is targeted by special ‘amyloid dyes’ like thioflavin T (ThT) or Congo red. Previously we had a particularly poor understanding of this part of the fibril surface, mostly because its NMR signals are easily overwhelmed by the much larger core that is sequestered from the surrounding water. The MD shows that this surface is much more dynamic than the fibril core, while it still maintains (on average) some of its structural features (in terms of the glutamine side chain configurations).
To also look at this experimentally, we thought of a way to see the surface through an isotope-editing approach: we can exchange key hydrogens (1H) in the fibril core with deuterium (2H), which makes them invisible in 1H-detected NMR. Then, we could back-exchange the surface with 1H hydrogens, and make the surface visible again, while the core remains invisible. (And vice versa, of course). These NMR measurements were performed in Utrecht, with the team of Markus Weingarth. The great complementarity of the MD and NMR approaches now gives us a new understanding of this part of the fibril surface. Aside from being bound by dyes like ThT, there is an interest in this surface for the design of PET ligands to be used in pre-clinical research, and perhaps ultimately clinical trials. (E.g in HTT lowering drug treatments it would be invaluable to be able to monitor the level of HTT inclusions during treatment)
In any case, we hope that our work can help the HD field in deepening our understanding of the life cycle of HTT and its fragments. Of course, there are also many open questions that remain. One obvious one is that these are proteins that aggregated in laboratory conditions. What happens in a real patient? Would these aggregates be similar or different? We discuss some considerations about this in the paper. Please see the paper for more details, and let us know if you have questions. (The code and coordinates are available on Zenodo)
Citation: Bagherpoor Helabad, M., Matlahov, I., Kumar, R. et al. Integrative determination of atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington disease. Nat Commun 15, 10793 (2024). https://doi.org/10.1038/s41467-024-55062-8
PDB coordinates of the HTTex1 fibril: download
Publication: NMR of 13C-labeled hyaluronic acid hydrogels (in context of ECM)
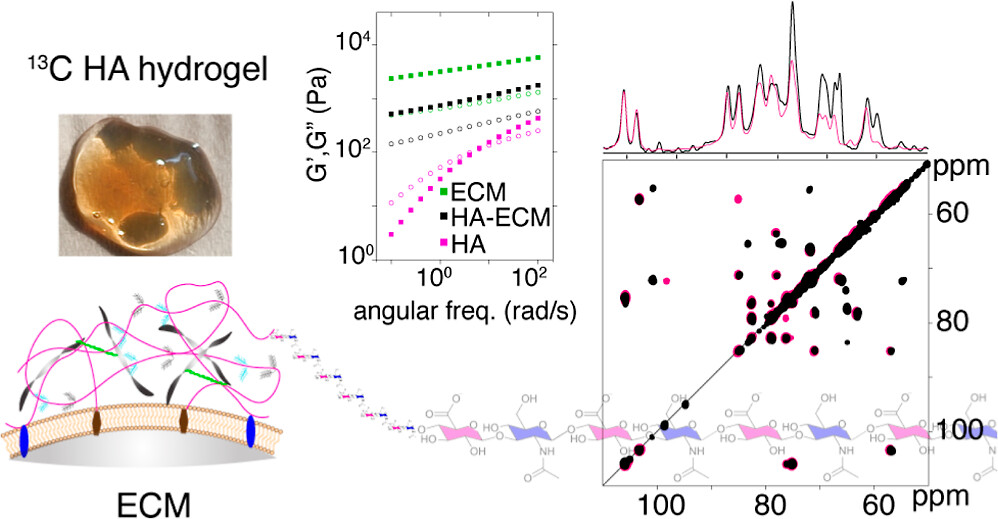
Congratulations to PhD student Pushpa Rampratap (and her co-authors!) on the publication of a new paper on the use of solid-state NMR spectroscopy to study hydrogels that mimic aspects of the extracellular matrix (ECM). Building on her previously published approach to produce 13C labeled hyaluronic acid (HA; with very high molecular weights), she performed extensive series of magic-angle-spinning NMR analyses of HA hydrogels under various conditions. Notably, this included ECM-mimicking conditions that are commonly used in cell culture and biomedical engineering studies (using the Geltrex ECM extract).
The resulting (very nice) paper shows the power of combining 13C enrichment with modern MAS NMR to gain truly atomic-level insights into the behavior of complex hydrogels (or ECMs). Surprisingly (to us), we observed highly localized changes affecting specific atoms in the HA, with the affected atoms being different from what we had expected. We briefly discuss the implications of this finding for e.g. HA-interacting proteins in a biological context. That said, these methods should be particularly powerful also for studying engeneering HA-based hydrogels and nanoparticles, which are finding widespread uses in different types of industries.
Citation:
Rampratap et al. (2024) Resolving Atomic-Level Dynamics and Interactions of High-Molecular-Weight Hyaluronic Acid by Multidimensional Solid-State NMR ACS Appl. Mater. Interfaces 2024, 16, 33, 43317–43328

Funding: new grant from the Barth Syndrome Foundation
We are excited to have received a Development Award grant from the international Barth Syndrome Foundation, enabling us to continue and develop our work studying the molecular mechanisms of this inherited lipid metabolism disease. This is in a sense a follow-up to our recent collaborative publication on this topic. In our paper in Nature Metabolism we used magic-angle-spinning NMR spectroscopy to probe a protein-lipid complex implicated in Barth Syndrome. The NMR analysis showed it to be highly dynamic, with enhanced mobility triggered by the lyso-cardiolipin lipids that are increased in BTHS. Moreover, we showed that it was feasible to probe small molecule interactions with this pathogenic protein-lipid complex, paving the way for further studies.
Some more information on the Foundation’s website. Or see our recent paper for some more background on the science behind the research. Related, earlier, research is also discussed on this page of our website.
Addendum: you can also find a video about the above mentioned paper, hosted by the Barth Syndrome Foundation. In it our co-authors explain and discuss the findings of that work and its relevance to the disease.
Visitor lecture series on solution-state NMR analysis
In the week of May 13-17, we will have an international visitor coming to present a series of educational and research lectures at the University of Groningen. Dr. Andrea Cesari will present a series of four lectures on various aspects of solution-state NMR analysis. Everyone is welcome to attend. For questions please contact the host P.C.A. van der Wel (p.c.a.van.der.wel@rug.nl).
Speaker Dr. Andrea Cesari
Address Fixed-term Junior Researcher (RTDa), Organic Chemist and NMR Spectroscopist at Università di Pisa
Lecture titles:
1. From Sample to Spectrum: good practises in liquid-state NMR Spectroscopy
2. Exploring the NMR toolkit for molecular recognition studies – from relaxation times to 2D maps
3. Applications of liquid-state NMR spectroscopy in pharmaceutical technology
4. Nanoparticle-based chemosensors using NMR spectroscopy
More info here
Publication: New paper about photochemical approaches to studying polyQ protein aggregation.
Congratulations to PhD student Raffaella Parlato, Dr. Jana Volaric, and their collaborators, on the publication of a nice new paper in the Journal of the American Chemical Society. This is the final result of an idea from some years ago, which came together very nicely thanks to a great team of collaborators. The goal of this work was to explore the idea of putting polyglutamine aggregation under some degree of photo-control. In prior work, it has been shown that b-hairpin formation is a key step in the aggregation process of expanded polyQ proteins. Azobenzene-based groups can be used to favor or disfavor beta turn structures under the influence of light, but prior ways of implementing this have been limited in various ways. A new amino acid analogue is introduced with more favorable structural and photochemical properties. And, we tested it in the context of polyQ peptides, seeing it indeed modulating the conformation of resulting aggregates (seen by ssNMR on unlabeled peptide fibrils!). For more details, see the paper. It is open access, so easily accessible.
Citation:
Raffaella Parlato, Jana Volarić, Alessia Lasorsa, Mahdi Bagherpoor Helabad, Piermichele Kobauri, Greeshma Jain, Markus S. Miettinen, Ben L. Feringa*, Wiktor Szymanski*, and Patrick C. A. van der Wel* (2024) J. Am. Chem. Soc. 2024, 146, 3, 2062–2071. Photocontrol of the β-Hairpin Polypeptide Structure through an Optimized Azobenzene-Based Amino Acid Analogue. DOI: https://doi.org/10.1021/jacs.3c11155