Author: Patrick

Publication: Dynamics-based spectral editing to see (fiber) surfaces by solid-state NMR.
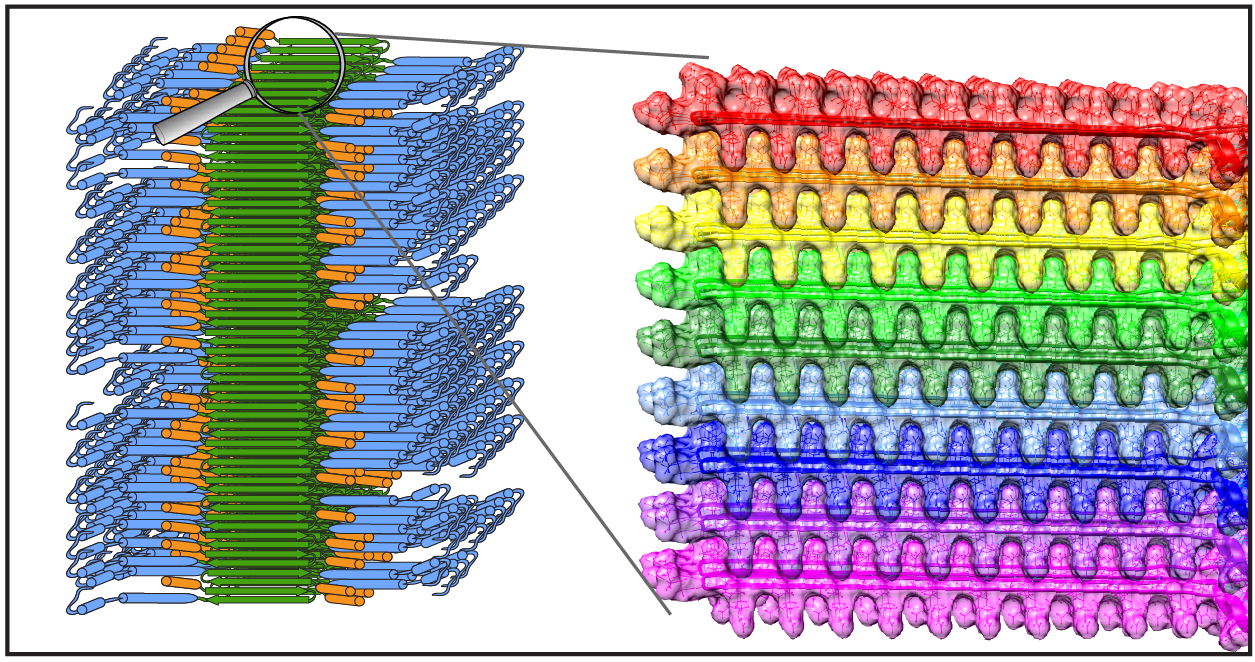
Congratulations to Dr. Irina Matlahov and (alum) Dr. Jennifer Boatz on the publication of their new paper in the Journal of Structural Biology X. The paper is entitled “Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils“. It describes our latest research on the misfolded protein deposits associated with Huntington’s disease (HD), specifically looking at what happens on the surface of these protein fibrils. In previous work we have studied the structure of these nanometer-sized fibrils formed by mutant huntingtin’s exon 1 fragment, using ssNMR, EM and other methods. In our earlier studies we used dynamics-sensitive […]

Publication: Collaborative paper with the Pescarmona group (ENTEG) on zeolites.
Congratulations to PhD student Mustapha El Hariri El Nokab and our collaborators from the Pescarmona group at ENTEG, on a new collaborative paper being accepted and posted online. In this work, Mustapha used both 29Si and 27Al magic angle spinning ssNMR to compare different zeolite samples, characterizing their chemical structure and degree of order. Aside from our ssNMR data, the paper features numerous other spectroscopic and synthetic methods. For more information, see the paper! Zahra Asgar Pour, Romar Koelewijn, Mustapha El Hariri El Nokab, Patrick C. A. van der Wel, Khaled O. Sebakhy, Paolo Pescarmona (2022) Binder-free zeolite Beta beads […]
News: Dutch-language radio
Recently our research was covered (briefly) on a Dutch radio station, featuring an interview with Patrick. It is mostly about our work applying ssNMR to studying how proteins aggregate in Huntington’s disease. You can find the recording online at the NPO radio website. More information about our Huntington disease research can also be found on this page, and in two recent webinars.

Upcoming event: BPS networking event on polyQ biophysics
Hereby an early announcement of an upcoming online event hosted by our group and the group of Markus Miettinen: “Biophysics of polyglutamine aggregation: how does it start and how does it end?“ This is an online event is organized in context of the networking event series sponsored and organized by the Biophysical Society (BPS). Our event aims to bring together international scientists working on the biophysics of polyglutamine (polyQ) protein aggregation. The event will take place on July 6th 2022, from 15:00-18:00 CET. More information can be found at the page from the BPS or our own institute. An online […]

Publication: SSNMR of alginate hydrogel (re)hydration
A new open-access publication by Mustapha and collaborators has been published in the journal Food Hydrocolloids, describing how he used various ssNMR measurements to probe alginate hydrogel structure and (re)hydration. Alginates can be cross-linked with calcium to form hydrogels, which are used in many different types of applications. This includes their use in slow-release drug delivery as well as food/nutritional applications. In this new paper, Mustapha shows how 1H, 2H and 13C ssNMR measurements can be used to see how these hydrogels are structured, but especially also how they are hydrated by the aqueous solvent. A key feature of the […]

Webinar: Huntington’s disease mechanisms.
On Sept 28 2021, Patrick gave a public webinar on the topic of protein Misfolding and aggregation in Huntington’s disease (and related polyglutamine disorders). This was part of the international webinar series on Cellular and Protein Homeostasis. This webinar is now available for streaming online at the following URL (updated URL in 2024). The webinar discusses some of the lessons we learned from our ongoing studies of what happens to mutated proteins in Huntington’s disease. In particular: what is the role of the mutated polyglutamine domain? How does it change its conformation as it transitions from its normal fold to […]

Publication: Structural and motional changes in a cytochrome c – lipid complex implicated in apoptosis.
Congratulations to lab alum Dr. Mingyue Li and our collaborators on the publication of a new paper in the Journal of Molecular Biology. The paper is online via its DOI link. The paper describes how we used solid-state NMR spectroscopy to characterise the partial destabilization of the native fold of the protein cytochrome c, as it is bound to cardiolipin lipids. This protein-lipid complex is implicated in the process of programmed cell death, where it plays a key role in triggering the self-destruction of undesired or disease cells in higher organisms. Notably, the CL-bound protein catalyses the process of mitochondrial […]

Publication: New paper on mitochondrial protein-lipid interactions published in PNAS.
Congratulations to lab alumns Dr. Abshishek Mandal and Dr. Jennifer Boatz, as well as our collaborators in the USA and Spain! A new collaborative paper on mitochondrial protein-lipid interactions has just been published in the journal PNAS. In this multidisciplinary work we studied how the protein Drp1 binds the special mitochondrial lipid cardiolipin in order to do its job managing the proper fission of mitochondrial membrane. Our collaborator Rajesh Ramachandran (at Case Western) coordinated a wide array of experimental and computational approaches to determine how Drp1’s “variable domain” (VD) binds cardiolipin. Along the way, two apparent CL-binding motifs were detected, […]

Webinar: Structural biology of Huntington’s disease.
On May 15th 2021, Patrick gave an invited lecture in the online webinar series on the Molecular Bases of Proteinopathies hosted by the Ramamoorthy lab at the University of Michigan. Patrick talked about our research on polyglutamine protein aggregation and the structural biology of Huntington’s disease.The seminar title was “The Structural Biology of Protein Misfolding in Huntington’s disease”. If you are interested, the seminar has been posted to YouTube, where it can be viewed here.

Preprint: cytochrome c – cardiolipin studies by ssNMR on bioRxiv
Our latest update on structural studies of this peroxidase-active protein-lipid complex implicated in mitochondrial apoptosis has now posted to bioRxiv: https://www.biorxiv.org/content/10.1101/2021.02.24.432556v1 In the preprint we discuss how ssNMR reveals the involvement of specific and localised dynamics in the lipid-bound protein. Interestingly, the mobility is dependent on the bound lipid species, with an apparent correlation to the resulting peroxidase activity. The lipids thus act as both substrates and regulators of the pro-apoptotic enzymatic activity of the protein. This was also discussed in the recent webinar as discussed in an earlier post. This work was made possible by several great collaborators at […]