Canonical NMR is optimized for studies of proteins and small molecules in solution. Nonetheless, solid or semi-solid samples play important roles in biology and biomedicine. We employ “solid-state” NMR to study the molecular structure and dynamics of such samples, in order to gain new insights into the mechanisms of life and disease.
Magic Angle Spinning (MAS) NMR
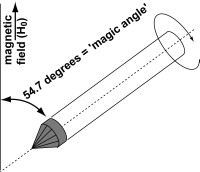
One approach is to emulate the motional avaraging effects that produce the narrow lines seen in solution NMR. This is accomplished by rapidly spinning the entire “solid” NMR sample in one particular orientation, with a 54.7 degree angle (the ‘magic angle‘) relative to the NMR spectrometer’s magnetic field. This approach is generally applicable to a wide range of samples, ranging from small molecule powders to large biomolecules. Examples of biological interest include nano- or microcrystalline proteins, membrane-bound proteins or peptide, as well as fibrillar and non-fibrillar protein deposits. These MAS NMR experiments can provide site-specific measurement of various structural constraints (atom-to-atom distances and torsion angles) as well as dynamics information (more here).
Oriented solid state NMR
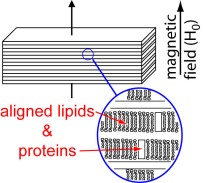
Aligning the material of interest in a single orientation (rather than the normal random distribution found in powder samples), also eliminates much of the broadening that is otherwise seen in solid samples. Lipid bilayers have a natural tendency to self-assemble in an aligned fashion, which allows structural measurements in an environment that closely resembles the biological membrane. This approach allows a detailed examination of protein-lipid interactions, for instance by using 2H GALA ssNMR experiments. Membrane alignment can be accomplished by deposition on glass slides (see right), or by using bicelles. These experiments provide unique insight into the orientation of membrane proteins as well as amino acid side chains.