Author: Patrick
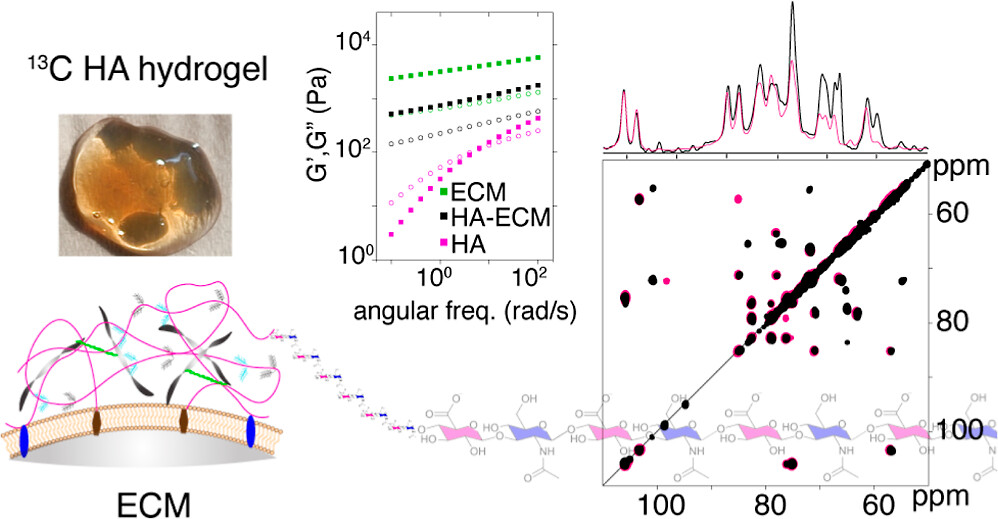
Publication: NMR of 13C-labeled hyaluronic acid hydrogels (in context of ECM)
Congratulations to PhD student Pushpa Rampratap (and her co-authors!) on the publication of a new paper on the use of solid-state NMR spectroscopy to study hydrogels that mimic aspects of the extracellular matrix (ECM). Building on her previously published approach to produce 13C labeled hyaluronic acid (HA; with very high molecular weights), she performed extensive series of magic-angle-spinning NMR analyses of HA hydrogels under various conditions. Notably, this included ECM-mimicking conditions that are commonly used in cell culture and biomedical engineering studies (using the Geltrex ECM extract). The resulting (very nice) paper shows the power of combining 13C enrichment with […]

Funding: new grant from the Barth Syndrome Foundation
We are excited to have received a Development Award grant from the international Barth Syndrome Foundation, enabling us to continue and develop our work studying the molecular mechanisms of this inherited lipid metabolism disease. This is in a sense a follow-up to our recent collaborative publication on this topic. In our paper in Nature Metabolism we used magic-angle-spinning NMR spectroscopy to probe a protein-lipid complex implicated in Barth Syndrome. The NMR analysis showed it to be highly dynamic, with enhanced mobility triggered by the lyso-cardiolipin lipids that are increased in BTHS. Moreover, we showed that it was feasible to probe […]

Visitor lecture series on solution-state NMR analysis
In the week of May 13-17, we will have an international visitor coming to present a series of educational and research lectures at the University of Groningen. Dr. Andrea Cesari will present a series of four lectures on various aspects of solution-state NMR analysis. Everyone is welcome to attend. For questions please contact the host P.C.A. van der Wel (p.c.a.van.der.wel@rug.nl). Speaker Dr. Andrea Cesari Address Fixed-term Junior Researcher (RTDa), Organic Chemist and NMR Spectroscopist at Università di Pisa Lecture titles: 1. From Sample to Spectrum: good practises in liquid-state NMR Spectroscopy 2. Exploring the NMR toolkit for molecular recognition studies – from relaxation times […]

Publication: New paper about photochemical approaches to studying polyQ protein aggregation.
Congratulations to PhD student Raffaella Parlato, Dr. Jana Volaric, and their collaborators, on the publication of a nice new paper in the Journal of the American Chemical Society. This is the final result of an idea from some years ago, which came together very nicely thanks to a great team of collaborators. The goal of this work was to explore the idea of putting polyglutamine aggregation under some degree of photo-control. In prior work, it has been shown that b-hairpin formation is a key step in the aggregation process of expanded polyQ proteins. Azobenzene-based groups can be used to favor […]

Publication: Review article on polyglutamine protein analysis by solid-state NMR spectroscopy.
Our new review article summarizing progress in polyglutamine protein studies by solid-state NMR is now available on the website of the journal Biochemical Society Transactions. This is an invited mini-review that was requested to cover the progress made in our understanding the aggregation and misfolding of mutant polyglutamine proteins implicated in diseases such as Huntington’s disease and several versions of spinocerebellar ataxia (SCA). We describe how studies from ssNMR has been used to better understand the structure of these protein aggregates, and thus helps us analyze how the aggregation process occurs and how aggregates may play a role in these […]

Publication: collaborative paper on perovskite-related materials, with Sn and Se ssNMR
Congrats to our collaborators from the group of Maria Loi and others for the publication of their new paper on in situ SnSe deposition as passivation for scalable and stable quasi-2D lead–tin perovskite solar cells, in the journal Energy & Environmental Science. Dr. Lasorsa in our group contributed tin and selenium ssNMR analysis to this interdisciplinary paper. For more information see the paper at the journal. Publication: Chen L, Tekelenburg EK, Gahlot K, Pitaro M, Xi J, Lasorsa A, et al. In situ SnSe deposition as passivation for scalable and stable quasi-2D lead–tin perovskite solar cells. Energy Environ Sci. 2023;10.1039.D3EE02507A.

Upcoming Symposium Dutch Protein Aggregation Network DPAN
Hold the date! On November 30th there will be an inaugural symposium of the Dutch Protein Aggregation Network (DPAN), sponsored by NWO. This one-day even (free registration) will be held at the VU in Amsterdam. It welcomes all Dutch researchers interested in protein aggregation related to human disease (including PIs, PhD students, postdocs..). The DPAN network aims to bring together the protein aggregation researchers of The Netherlands. With the help of NWO sponsorship DPAN will start with a one-day symposium on Nov 30th. This will take place at the O|2 building of the VU in Amsterdam. The keynote speaker will […]

Educational webinar on the use of ssNMR to measure dihedral angles.
Recently, Patrick gave an online lecture in the online webinar tutorial series of the Global NMR Discussion Meetings series (episode 70!). In this online lecture he introduced and discussed approaches to measure torsion angles (or dihedral angles) using advanced solid-state NMR spectroscopy. He discussed the basic idea of how these are implemented in ssNMR, but also how such structural data can be a useful complement to more traditional inter-atomic distance information obtained by ssNMR. You can now watch the recording of this lecture on Youtube, via this link. Want to learn even more? This lecture is connected to our recent […]

Publication: 13C labeling of hyaluronic acid polysaccharides (for ssNMR analysis)
Congrats to Pushpa and her collaborators on the new paper in the journal Carbohydrate Polymers. This new report describes Pushpa’s work to achieve the production of high-molecular-weight (HMW) hyaluronic acid, as part of her PhyCan (physics of cancer) project. Our interest in this polysaccharide stems from its important role in the extracellular matrix (ECM) of tissues, and in particular certain types of cancer tissues. In such tumors the amount of HA is upregulated, seemingly contributing to the progression of cancer development. A challenge in understanding this process is that HMW is difficult to study with most structural techniques, as it […]

Job search: NMR spectroscopist being recruited at the RuG
The University of Groningen is looking to hire a NMR spectroscopist for the solution NMR facility associated with the chemistry-focused Stratingh Institute. Ad is here: https://www.rug.nl/about-ug/work-with-us/job-opportunities/?details=00347-02S0009SXP&cat=obp Note that this position is not associated with our research group. The mentioned NMR facility is part of the Stratingh Institute and focuses on solution NMR studies.