Congratulations to Irina, Greeshma, and Raffaella, together with our international network of collaborators, on a newly published paper that came out in Nature Communications in Dec 2024, in a special collection on the Biology of rare genetic disorders. In this collaborative work, we looked at the structure of proteins behind Huntington’s disease (HD), enabled by crucial funding from the CampagneTeam Huntington and the CHDI Foundation. HD is an inherited neurodegenerative disease, with patients having a mutated form of the huntingtin (HTT) gene. The mutation affects a repeating sequence of CAG codons that has been expanded in HD patients. This translates to huntingtin (HTT) proteins featuring an expanded polyglutamine (or polyQ) segment. Most people have about 20 residues in their HTT polyglutamine segment. Patients have more than 35. Postmortem analysis of HD patient brains reveals the presence of protein clumps (aggregates or inclusions) that contain short fragments of the mutated HTT protein.
In past work we (and others) have studied how HTT fragments also form aggregates when you make the proteins in the lab, and do so more and more aggressively as you expand their polyglutamine segments. Although the precise role of these protein clumps in the disease remains under active debate, there is a consensus that they represent an important biomarker of the disease. Until now, we have lacked a solid understanding of the molecular conformation of the HTT protein in these clumps. For some years now my group (with various collaborators) has been trying to address this question.
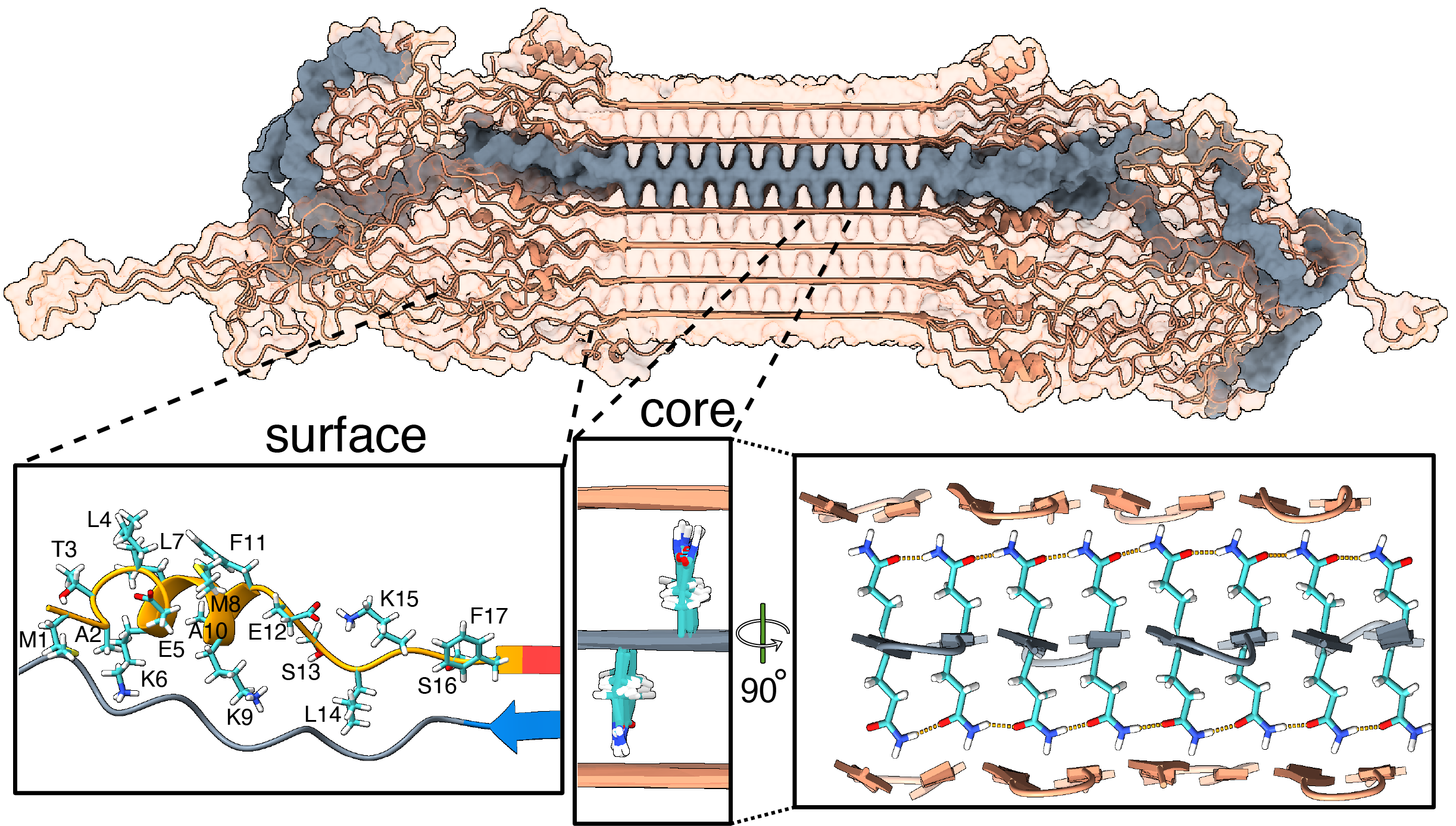
Our new paper (together with the team of Markus Miettinen, University of Bergen) introduces the first atomic-level description of the HTT fragments that are increasingly thought to be relevant to the disease. We worked closely together to combine ssNMR data with results from other techniques to build a comprehensive and integrative model that visualizes these protein fibrils in atomic detail. Key aspects of the resulting structure are of course informed and determined by our experimental findings. However, the modeling also unveiled important and useful new insights.
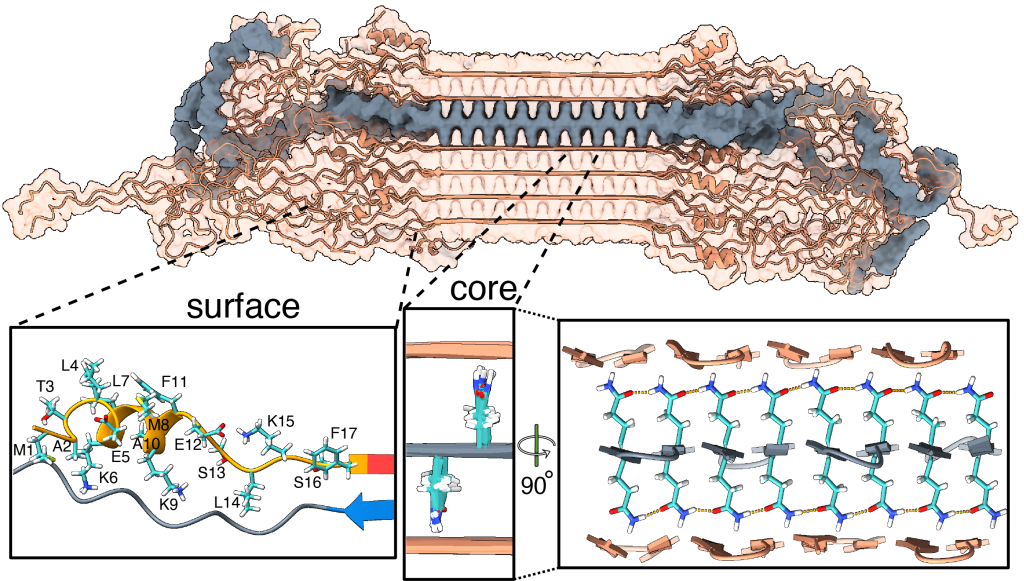
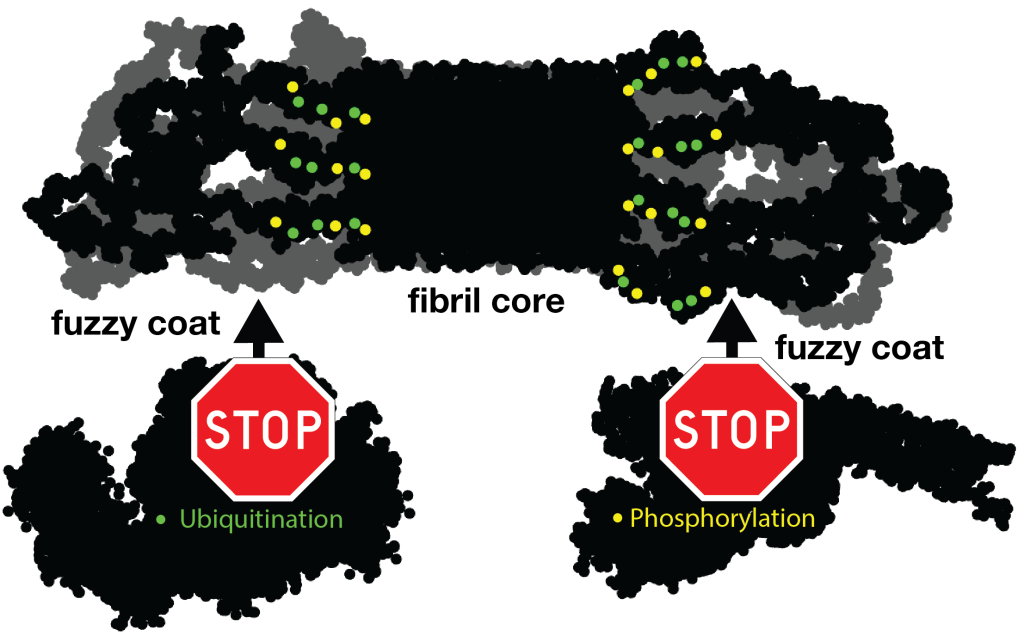
For instance, let’s look at the fuzzy coat that covers these HD amyloid fibrils. Regions outside the mutated polyQ stretch (known as ‘flanking segments’) form a disordered, but densely packed, brush on two sides of a roughly rectangular fibril core. We discuss in the paper how the dense packing reduces or prohibits interactions of other proteins with the flanking segments. This limits the ability of kinases to phosphorylate the aggregated proteins; and similarly for poly-ubiquitination. This may be seen as bad news, as both processes may be used to enable destabilization or destruction of the aggregates.
However, these brushes cover only two sides of the fibril core. The other two sides feature an extensive interface directly between the polyQ amyloid core and the surrounding aqueous phase. In laboratory experiments, it is this type of interface that is targeted by special ‘amyloid dyes’ like thioflavin T (ThT) or Congo red. Previously we had a particularly poor understanding of this part of the fibril surface, mostly because its NMR signals are easily overwhelmed by the much larger core that is sequestered from the surrounding water. The MD shows that this surface is much more dynamic than the fibril core, while it still maintains (on average) some of its structural features (in terms of the glutamine side chain configurations).
To also look at this experimentally, we thought of a way to see the surface through an isotope-editing approach: we can exchange key hydrogens (1H) in the fibril core with deuterium (2H), which makes them invisible in 1H-detected NMR. Then, we could back-exchange the surface with 1H hydrogens, and make the surface visible again, while the core remains invisible. (And vice versa, of course). These NMR measurements were performed in Utrecht, with the team of Markus Weingarth. The great complementarity of the MD and NMR approaches now gives us a new understanding of this part of the fibril surface. Aside from being bound by dyes like ThT, there is an interest in this surface for the design of PET ligands to be used in pre-clinical research, and perhaps ultimately clinical trials. (E.g., in HTT lowering drug treatments it would be invaluable to be able to monitor the level of HTT inclusions during treatment)
In any case, we hope that our work can help the HD field in deepening our understanding of the life cycle of HTT and its fragments. Of course, there are also many open questions that remain. One obvious one is that these are proteins that aggregated in laboratory conditions. What happens in a real patient? Would these aggregates be similar or different? We discuss some considerations about this in the paper. Please see the paper for more details, and let us know if you have questions. (The code and coordinates are available on Zenodo)
Citation: Bagherpoor Helabad, M., Matlahov, I., Kumar, R. et al. Integrative determination of atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington disease. Nat Commun 15, 10793 (2024). https://doi.org/10.1038/s41467-024-55062-8. (Part of the special collection Biology of rare genetic disorders)
PDB coordinates of the HTTex1 fibril: download
NB. this article was highlighted in the HD-Buzz blog in August 2025.