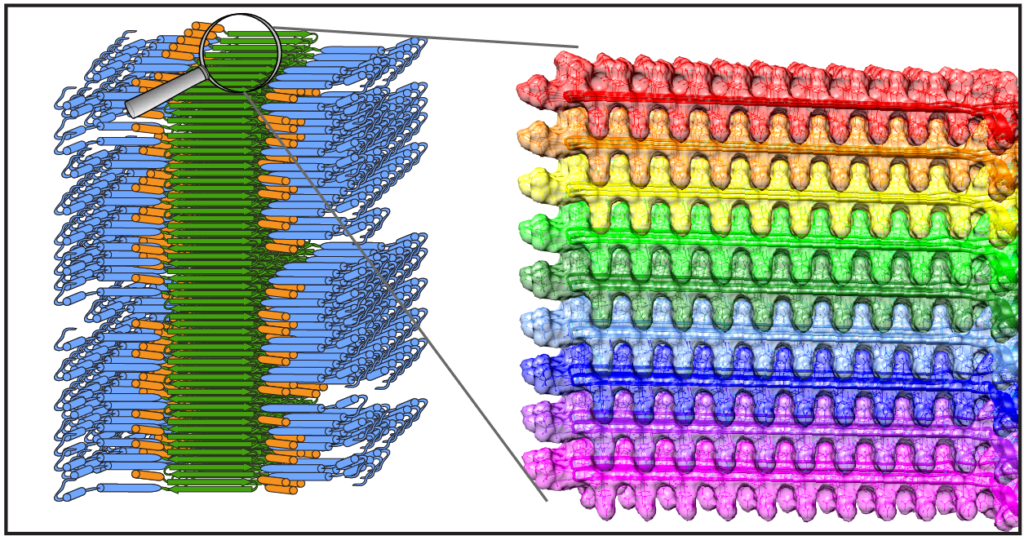
Congratulations to Dr. Jennifer Boatz and other team members for the acceptance and publication of a nice new paper on the structure of the misfolded mutant protein from Huntington’s disease. The paper is online at the Journal of Molecular Biology. Based on an integration of multiple techniques (NMR, EM, and X-ray diffraction), Jennifer assembled a new structural model of the protofilaments that make up the hierarchical fiber architecture of mutant huntingtin exon 1. This is a timely and important step forward in our understanding of the (mis)behaviour of the HD protein, and in particular how it forms pathogenic inclusions and protein aggregates.
In this new paper and prior work we (and others) have seen that the mutant protein is prone to form a collection of different kinds of aggregates, with each their own structure and functional properties. This is of substantial interest from a disease perspective, as these “functional properties” can encompass different degrees of neurotoxic properties. Surprisingly, Jennifer shows in this paper that one contributor to, or trigger of, huntingtin polymorphism is the concentration of the protein. Further studies will have to explore whether or how this finding impacts efforts to replicate cellular behaviour of the protein in vitro, and how it may affect the toxic properties of the aggregated protein states.
Reference:
Protofilament Structure and Supramolecular Polymorphism of Aggregated Mutant Huntingtin Exon 1. Boatz, J.C., Piretra, T., Lasorsa, A., Matlahov, I., Conway, J.F. & Van der Wel, P.C.A. (2020) J. Mol. Biol., 432(16): 4722-4744. [DOI] (Open Access)
Funding & support: The underlying research in this paper was enabled by funding support from the American NIH/NIGMS (grant R01 GM112678) and funding from the CampagneTeam Huntington in the Netherlands. For more information on the disease, see also our HD page and the CTH website. Other support came from the University of Groningen and the University of Pittsburgh.
Note added: Our Institute also highlighted this paper in a nice summary posted on the Zernike Institute website.