Blog Posts
Publication: Review article on polyglutamine protein analysis by solid-state NMR spectroscopy.
Our new review article summarizing progress in polyglutamine protein studies by solid-state NMR is now available on the website of the journal Biochemical Society Transactions. This is an invited mini-review that was requested to cover the progress made in our understanding the aggregation and misfolding of mutant polyglutamine proteins implicated in diseases such as Huntington’s disease and several versions of spinocerebellar ataxia (SCA). We describe how studies from ssNMR has been used to better understand the structure of these protein aggregates, and thus helps us analyze how the aggregation process occurs and how aggregates may play a role in these diseases. The paper is published open-access, so you should be able to read it to see all the details. (Naturally, the format of the mini-review limited the word count and meant that much had to be omitted)
Citation:
Patrick C.A. van der Wel; Solid-state nuclear magnetic resonance in the structural study of polyglutamine aggregation. Biochem Soc Trans 2024; BST20230731. doi: https://doi.org/10.1042/BST20230731
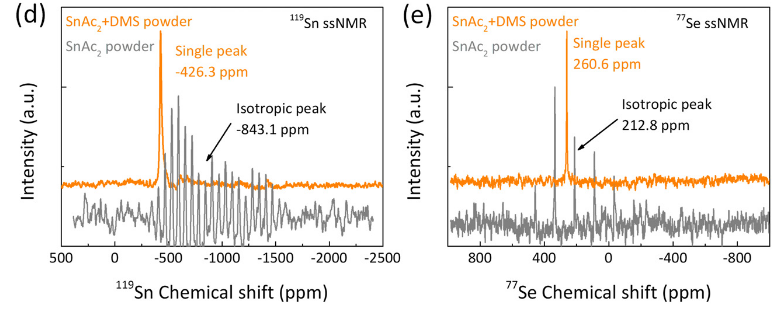
Publication: collaborative paper on perovskite-related materials, with Sn and Se ssNMR
Congrats to our collaborators from the group of Maria Loi and others for the publication of their new paper on in situ SnSe deposition as passivation for scalable and stable quasi-2D lead–tin perovskite solar cells, in the journal Energy & Environmental Science. Dr. Lasorsa in our group contributed tin and selenium ssNMR analysis to this interdisciplinary paper. For more information see the paper at the journal.
Publication:
Chen L, Tekelenburg EK, Gahlot K, Pitaro M, Xi J, Lasorsa A, et al. In situ SnSe deposition as passivation for scalable and stable quasi-2D lead–tin perovskite solar cells. Energy Environ Sci. 2023;10.1039.D3EE02507A.
Educational webinar on the use of ssNMR to measure dihedral angles.
Recently, Patrick gave an online lecture in the online webinar tutorial series of the Global NMR Discussion Meetings series (episode 70!). In this online lecture he introduced and discussed approaches to measure torsion angles (or dihedral angles) using advanced solid-state NMR spectroscopy. He discussed the basic idea of how these are implemented in ssNMR, but also how such structural data can be a useful complement to more traditional inter-atomic distance information obtained by ssNMR. You can now watch the recording of this lecture on Youtube, via this link.
Want to learn even more?
This lecture is connected to our recent review article on the same topic, which can be found online at: Frontiers | Dihedral Angle Measurements for Structure Determination by Biomolecular Solid-State NMR Spectroscopy (frontiersin.org)
Publication: Dynamics-based spectral editing to see (fiber) surfaces by solid-state NMR.
Congratulations to Dr. Irina Matlahov and (alum) Dr. Jennifer Boatz on the publication of their new paper in the Journal of Structural Biology X. The paper is entitled “Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils“. It describes our latest research on the misfolded protein deposits associated with Huntington’s disease (HD), specifically looking at what happens on the surface of these protein fibrils.
In previous work we have studied the structure of these nanometer-sized fibrils formed by mutant huntingtin’s exon 1 fragment, using ssNMR, EM and other methods. In our earlier studies we used dynamics-sensitive NMR techniques to look at the core of these fibrils, taking advantage of its very rigid structure. Conversely, we had looked at super flexible protein parts that are away from the rigid core. Those types of techniques have been a popular tool for seeing rigid or flexible parts of (aggregated) proteins. However, in the current paper we try to look for protein parts with intermediate mobility. Of particular interest are those residues that form the surface of the rigid fibril core: residues that are somewhat immobilized by proximity to the core proper, but are somewhat mobilized by their interactions with water solvent surrounding the fibril. This part of the protein fibrils can be especially interesting, as it is the part seen by protein-targeting antibodies, PET ligands and protein-protein interaction partners in affected neurons.
The paper discusses how we can probe the impact of the water interactions with the surface through variable temperature ssNMR. However, it also shows the limitations of this traditional approach. Instead we advocate for a new type of dynamic filtering experiment that selectively shows the signals of semi-rigid/semi-mobile residues (such as those on the fiber surface). This technique (IMS-DYSE) is complementary to traditional DYSE methods that select rigid or flexible sites. For more details, please see the paper (linked below). We expect to combine this dynamic filtering technique with other types of pulse sequences, to probe in more detail the features of the fiber surface.
Note that this technique proved especially useful for our work on polyglutamine protein fibrils, as in this protein system we have a core made up of glutamine residues, providing an overwhelming strong NMR signal that masks the resonances of the surface glutamines. So, to see the surface, it is essential to find a way to suppress the core signals, leaving only those of the surface residues. The paper shows that this indeed works, and we can for the first time use this technique to distinguish the core and surface glutamines. With this technique in hand, we can foresee further studies of surface-specific features, such as the binding sites for targeted antibodies and/or amyloid-binding dyes.
The paper is available online at the journal, and has the following citation:
I. Matlahov, J.C. Boatz, P.C.A. van der Wel (2022) Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils. J. Struct. Biol. X, vol. 6, 100077. DOI: 10.1016/j.yjsbx.2022.100077
PS. Some of the techniques and results from this publication were also discussed during a prior online seminar in the MIT ssNMR/DNP zoominar series. You can view that video here.
News: Dutch-language radio
Recently our research was covered (briefly) on a Dutch radio station, featuring an interview with Patrick. It is mostly about our work applying ssNMR to studying how proteins aggregate in Huntington’s disease. You can find the recording online at the NPO radio website. More information about our Huntington disease research can also be found on this page, and in two recent webinars.

Publication: SSNMR of alginate hydrogel (re)hydration
A new open-access publication by Mustapha and collaborators has been published in the journal Food Hydrocolloids, describing how he used various ssNMR measurements to probe alginate hydrogel structure and (re)hydration. Alginates can be cross-linked with calcium to form hydrogels, which are used in many different types of applications. This includes their use in slow-release drug delivery as well as food/nutritional applications. In this new paper, Mustapha shows how 1H, 2H and 13C ssNMR measurements can be used to see how these hydrogels are structured, but especially also how they are hydrated by the aqueous solvent. A key feature of the work is the use of 1H MAS NMR to detect the water molecules in the hydrogel macropores as well as those waters directly interacting with the alginate. The two types of water molecules form distinct ‘pools’ in the hydrogel, which can be nicely distinguished by their different molecular motion. The latter is detected via different relaxation measurements, in these NMR experiments.
To read more:
El Hariri El Nokab, M..; Lasorsa, A.; Sebakhy, K. O.; Picchioni, F.; van der Wel, P. C. A. Solid-State NMR Spectroscopy Insights for Resolving Different Water Pools in Alginate Hydrogels. Food Hydrocolloids 2022, 107500.
Publication: New paper on mitochondrial protein-lipid interactions published in PNAS.
Congratulations to lab alumns Dr. Abshishek Mandal and Dr. Jennifer Boatz, as well as our collaborators in the USA and Spain! A new collaborative paper on mitochondrial protein-lipid interactions has just been published in the journal PNAS. In this multidisciplinary work we studied how the protein Drp1 binds the special mitochondrial lipid cardiolipin in order to do its job managing the proper fission of mitochondrial membrane. Our collaborator Rajesh Ramachandran (at Case Western) coordinated a wide array of experimental and computational approaches to determine how Drp1’s “variable domain” (VD) binds cardiolipin. Along the way, two apparent CL-binding motifs were detected, which are seemingly shared by other CL-binding proteins. Their mutation disrupts CL binding and also the proper management of mitochondrial morphology, pointing to the importance of CL-based signals and interactions in this vital cellular process.
Experimentally, our group contributed various solid-state NMR measurements of the lipids, with which we probe the specificity of the interactions and also observe how the protein modulates the lipid bilayer itself. Moreover, we were involved in the sequence/structure analysis: sequence analysis of the “disordered” VD domain pinpointed those parts of the structure most likely to engage in lipid-driven folding upon membrane binding. The “MoRF” motifs (“molecular recognition features”) indeed appear to be involved in CL binding, based on NMR and mutational studies. In Drp1 they are intimately involved in a folding transition of the VD domain upon membrane binding. It will be interesting to see how widespread these CL-binding motifs (CBMs) are in other CL-binding proteins.
Reference:
[1] Mahajan M, Bharambe N, Shang Y, Lu B, Mandal A, Madan Mohan P, et al. NMR identification of a conserved Drp1 cardiolipin-binding motif essential for stress-induced mitochondrial fission. Proc Natl Acad Sci USA2021;118:e2023079118. https://doi.org/10.1073/pnas.2023079118.
New (e)book on membrane studies by solid-state NMR released online.
Now available online: we have contributed a chapter to a new (e)book on the topic of solid-state NMR studies of membranes and membrane proteins, edited by Frances Separovic and Marc-Antoine Sani (Univ. Melbourne, Australia). The edited volume “Solid state NMR. Applications in biomembrane structure.” was released in the IOP series in association with the Biophysical Society.
Our chapter (Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions [1]) looks at several studies that use ssNMR to probe peripheral membrane proteins. A key focus is on our own work on the mitochondrial protein cytochrome c, and how it binds to cardiolipin lipids during apoptosis, funded by the NIH/NIGMS [2]. In the chapter we try to summarise some of the practical challenges involved, along with potential solutions reported by ourselves and a few other research groups that studied other peripheral membrane proteins by ssNMR.
Cited references:
[1] Van der Wel, P.C.A. (2020) Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions. Chapter 10 in Solid-state NMR; applications in biomembrane structure. Edited by F. Separovic & M.-A. Sani; IOP Press (DOI 10.1088/978-0-7503-2532-5ch10)
[2] Li, M.; Mandal, A.; Tyurin, V. A.; DeLucia, M.; Ahn, J.; Kagan, V. E.; van der Wel, P. C. A. (2019) Surface-Binding to Cardiolipin Nanodomains Triggers Cytochrome c Pro-Apoptotic Peroxidase Activity via Localized Dynamics. Structure 2019, 27 (5), 806-815.e4. (DOI 10.1016/j.str.2019.02.007)
New publication on structure of mutant huntingtin protein from Huntington’s Disease
Congratulations to Dr. Jennifer Boatz and other team members for the acceptance and publication of a nice new paper on the structure of the misfolded mutant protein from Huntington’s disease. The paper is online at the Journal of Molecular Biology. Based on an integration of multiple techniques (NMR, EM, and X-ray diffraction), Jennifer assembled a new structural model of the protofilaments that make up the hierarchical fiber architecture of mutant huntingtin exon 1. This is a timely and important step forward in our understanding of the (mis)behaviour of the HD protein, and in particular how it forms pathogenic inclusions and protein aggregates.
In this new paper and prior work we (and others) have seen that the mutant protein is prone to form a collection of different kinds of aggregates, with each their own structure and functional properties. This is of substantial interest from a disease perspective, as these “functional properties” can encompass different degrees of neurotoxic properties. Surprisingly, Jennifer shows in this paper that one contributor to, or trigger of, huntingtin polymorphism is the concentration of the protein. Further studies will have to explore whether or how this finding impacts efforts to replicate cellular behaviour of the protein in vitro, and how it may affect the toxic properties of the aggregated protein states.
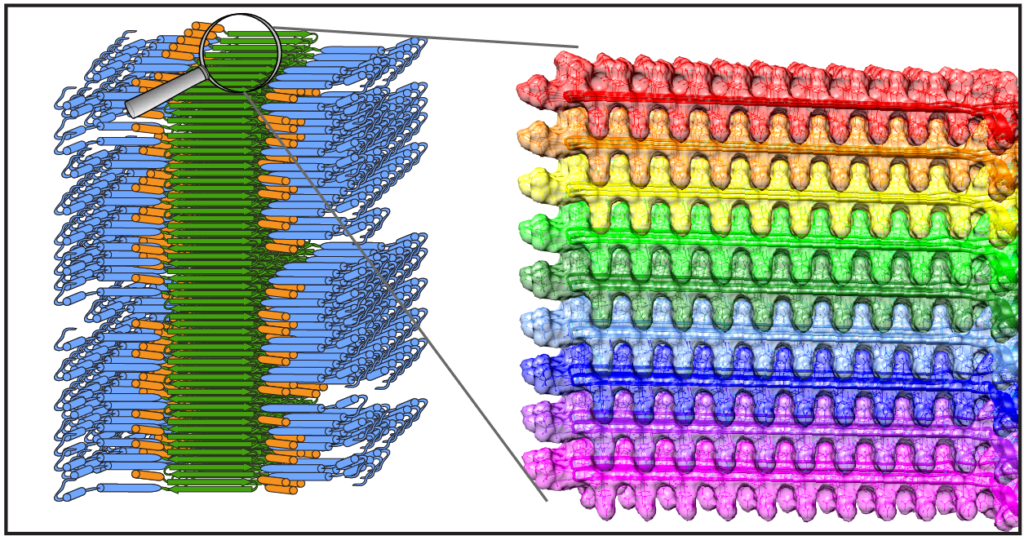
Reference:
Protofilament Structure and Supramolecular Polymorphism of Aggregated Mutant Huntingtin Exon 1. Boatz, J.C., Piretra, T., Lasorsa, A., Matlahov, I., Conway, J.F. & Van der Wel, P.C.A. (2020) J. Mol. Biol., 432(16): 4722-4744. [DOI] (Open Access)
Funding & support: The underlying research in this paper was enabled by funding support from the American NIH/NIGMS (grant R01 GM112678) and funding from the CampagneTeam Huntington in the Netherlands. For more information on the disease, see also our HD page and the CTH website. Other support came from the University of Groningen and the University of Pittsburgh.
Note added: Our Institute also highlighted this paper in a nice summary posted on the Zernike Institute website.
Competitive PhD Scholarships call open (deadline April 1st)
Our Institute and Faculty have opened up a competitive call for PhD scholarship applications. Our solid-state NMR group participates in one of the research theme areas, designated as “Advanced Materials”, which spans topics from physics of life, via bio-inspired materials, to materials and much more. The process is described in some detail on the RuG website. Briefly, applicants (with a MSc degree) are expected to contact a PI/supervisor (immediately!) and develop a fitting project to submit. The submission deadline of an initial idea (few hundred words) is due by April 1st 2020!
Suitable ideas are expected to fit within designated topics areas, described on the website here and here. Note especially also the 2nd link, as it contains important detailed information.T
Interested in this? Please contact us as soon as possible, to meet the tight deadlines. Together we can consider topics that range from self-assembling bio-inspired materials, Physics of Cancer, non-biological materials and other topics. Projects will be highly interdisciplinary as the involvement of a second supervisor is also required, with a distinct and complementary expertise.
Visitor lecture series on solution-state NMR analysis
In the week of May 13-17, we will have an international visitor coming to present a series of educational and research lectures at the University of Groningen. Dr. Andrea Cesari will present a series of four lectures on various aspects of solution-state NMR analysis. Everyone is welcome to attend. For questions please contact the host P.C.A. van der Wel (p.c.a.van.der.wel@rug.nl).
Speaker Dr. Andrea Cesari
Address Fixed-term Junior Researcher (RTDa), Organic Chemist and NMR Spectroscopist at Università di Pisa
Lecture titles:
1. From Sample to Spectrum: good practises in liquid-state NMR Spectroscopy
2. Exploring the NMR toolkit for molecular recognition studies – from relaxation times to 2D maps
3. Applications of liquid-state NMR spectroscopy in pharmaceutical technology
4. Nanoparticle-based chemosensors using NMR spectroscopy
More info here
Publication: New paper about photochemical approaches to studying polyQ protein aggregation.
Congratulations to PhD student Raffaella Parlato, Dr. Jana Volaric, and their collaborators, on the publication of a nice new paper in the Journal of the American Chemical Society. This is the final result of an idea from some years ago, which came together very nicely thanks to a great team of collaborators. The goal of this work was to explore the idea of putting polyglutamine aggregation under some degree of photo-control. In prior work, it has been shown that b-hairpin formation is a key step in the aggregation process of expanded polyQ proteins. Azobenzene-based groups can be used to favor or disfavor beta turn structures under the influence of light, but prior ways of implementing this have been limited in various ways. A new amino acid analogue is introduced with more favorable structural and photochemical properties. And, we tested it in the context of polyQ peptides, seeing it indeed modulating the conformation of resulting aggregates (seen by ssNMR on unlabeled peptide fibrils!). For more details, see the paper. It is open access, so easily accessible.
Citation:
Raffaella Parlato, Jana Volarić, Alessia Lasorsa, Mahdi Bagherpoor Helabad, Piermichele Kobauri, Greeshma Jain, Markus S. Miettinen, Ben L. Feringa*, Wiktor Szymanski*, and Patrick C. A. van der Wel* (2024) J. Am. Chem. Soc. 2024, 146, 3, 2062–2071. Photocontrol of the β-Hairpin Polypeptide Structure through an Optimized Azobenzene-Based Amino Acid Analogue. DOI: https://doi.org/10.1021/jacs.3c11155
Upcoming Symposium Dutch Protein Aggregation Network DPAN
Hold the date! On November 30th there will be an inaugural symposium of the Dutch Protein Aggregation Network (DPAN), sponsored by NWO. This one-day even (free registration) will be held at the VU in Amsterdam. It welcomes all Dutch researchers interested in protein aggregation related to human disease (including PIs, PhD students, postdocs..).
The DPAN network aims to bring together the protein aggregation researchers of The Netherlands. With the help of NWO sponsorship DPAN will start with a one-day symposium on Nov 30th. This will take place at the O|2 building of the VU in Amsterdam.
The keynote speaker will be Harm Kampinga from the UMCG Groningen. Many other speakers will discuss their research on amyloid formation and other aspects of protein aggregation.
Please join us and register here.
Publication: 13C labeling of hyaluronic acid polysaccharides (for ssNMR analysis)
Congrats to Pushpa and her collaborators on the new paper in the journal Carbohydrate Polymers. This new report describes Pushpa’s work to achieve the production of high-molecular-weight (HMW) hyaluronic acid, as part of her PhyCan (physics of cancer) project. Our interest in this polysaccharide stems from its important role in the extracellular matrix (ECM) of tissues, and in particular certain types of cancer tissues. In such tumors the amount of HA is upregulated, seemingly contributing to the progression of cancer development. A challenge in understanding this process is that HMW is difficult to study with most structural techniques, as it can be very large (megadaltons!) and highly dynamic. In this project, Pushpa outfitted HMW HA with 13C (and 15N) isotope labels, and shows that this makes multidimensional NMR (including solid-state NMR) feasible. This approach is expected to be valuable for many research areas, as HA is also important for many biomedical engineering (BME) type applications. ECM-mimicking hydrogels are for instance of great interest for engineering 3D environments for growing cells.
Publication:
Rampratap P, Lasorsa A, Perrone B, Van Der Wel PCA, Walvoort MTC. Production of isotopically enriched high molecular weight hyaluronic acid and characterization by solid-state NMR. Carbohydrate Polymers. 2023 Sep;316:121063.
Job search: NMR spectroscopist being recruited at the RuG
The University of Groningen is looking to hire a NMR spectroscopist for the solution NMR facility associated with the chemistry-focused Stratingh Institute. Ad is here: https://www.rug.nl/about-ug/work-with-us/job-opportunities/?details=00347-02S0009SXP&cat=obp
Note that this position is not associated with our research group. The mentioned NMR facility is part of the Stratingh Institute and focuses on solution NMR studies.
Publication: Collaborative paper with the Pescarmona group (ENTEG) on zeolites.
Congratulations to PhD student Mustapha El Hariri El Nokab and our collaborators from the Pescarmona group at ENTEG, on a new collaborative paper being accepted and posted online. In this work, Mustapha used both 29Si and 27Al magic angle spinning ssNMR to compare different zeolite samples, characterizing their chemical structure and degree of order. Aside from our ssNMR data, the paper features numerous other spectroscopic and synthetic methods. For more information, see the paper!
Zahra Asgar Pour, Romar Koelewijn, Mustapha El Hariri El Nokab, Patrick C. A. van der Wel, Khaled O. Sebakhy, Paolo Pescarmona (2022) Binder-free zeolite Beta beads with hierarchical porosity: synthesis and application as heterogeneous catalysts for anisole acylation. ChemCatChem in press.