Author: Patrick
MOSBRI Biophysics Infrastructure
Starting later this year, the RUG ssNMR group will be of the newly funded MOlecular-Scale Biophysics Research Infrastructure (MOSBRI) network. This EU-wide consortium enables ambitious integrative multi-technological studies of biological systems at the crucial intermediate level between atomic-resolution structural descriptions and cellular-scale observations. MOSBRI provides European academic and industrial researchers with a one-stop shop Trans-National Access to the latest technological developments in advanced spectroscopies, hydrodynamics, thermodynamics, real-time kinetics and single molecule approaches.
More information can be found on the MOSBRI website, and also on our own MOSBRI page.
This infrastructure network is supported by EU funding and features resources in many EU countries (and the UK). The page of the University of Groningen hub is found here.

Online presentation: our work featured in the Polymer Physics & Polymer Spectroscopy (P3S) webinar series
Patrick was invited to present some of our work in the P3S webinar series organised by three research groups from Europe, China and the US. This recurring series has included many interesting talks on studies of polymers and hydrogels by various spectroscopic techniques, with a recurring role for (solid-state) NMR spectroscopy. Patrick spoke about our published work on studying the repeating polymer structures of polyglutamine proteins [1] and (briefly) our newer investigations of polysaccharide hydrogels [2] . In the former topic, the talk discussed the use and benefits of torsion angle measurements by solid-state NMR, as demonstrated in our prior work on various different samples [1,3-4].
The talk was recorded and can be found in the P3S online archive, with the specific talk linked here.
- Hoop, C. L.; Lin, H.-K.; Kar, K.; Magyarfalvi, G.; Lamley, J. M.; Boatz, J. C.; Mandal, A.; Lewandowski, J. R.; Wetzel, R.; van der Wel, P. C. A. Huntingtin Exon 1 Fibrils Feature an Interdigitated β-Hairpin-Based Polyglutamine Core. Proc. Natl. Acad. Sci. USA 2016, 113 (6), 1546–1551. https://doi.org/10.1073/pnas.1521933113.
- El Hariri El Nokab, M.; van der Wel, P. C. A. Use of Solid-State NMR Spectroscopy for Investigating Polysaccharide-Based Hydrogels: A Review. Carbohydrate Polymers 2020, 116276. https://doi.org/10.1016/j.carbpol.2020.116276.
- Bajaj, V. S.; van der Wel, P. C. A.; Griffin, R. G. Observation of a Low-Temperature, Dynamically Driven Structural Transition in a Polypeptide by Solid-State NMR Spectroscopy. J Am Chem Soc 2009, 131 (1), 118–128. https://doi.org/10.1021/ja8045926.
- Van der Wel, P. C. A.; Lewandowski, J. R.; Griffin, R. G. Structural Characterization of GNNQQNY Amyloid Fibrils by Magic Angle Spinning NMR. Biochemistry 2010, 49 (44), 9457–9469. https://doi.org/10.1021/bi100077x.
New (e)book on membrane studies by solid-state NMR released online.
Now available online: we have contributed a chapter to a new (e)book on the topic of solid-state NMR studies of membranes and membrane proteins, edited by Frances Separovic and Marc-Antoine Sani (Univ. Melbourne, Australia). The edited volume “Solid state NMR. Applications in biomembrane structure.” was released in the IOP series in association with the Biophysical Society.
Our chapter (Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions [1]) looks at several studies that use ssNMR to probe peripheral membrane proteins. A key focus is on our own work on the mitochondrial protein cytochrome c, and how it binds to cardiolipin lipids during apoptosis, funded by the NIH/NIGMS [2]. In the chapter we try to summarise some of the practical challenges involved, along with potential solutions reported by ourselves and a few other research groups that studied other peripheral membrane proteins by ssNMR.
Cited references:
[1] Van der Wel, P.C.A. (2020) Solid-state NMR studies of peripherally membrane-associated proteins: dealing with dynamics, disorder and dilute conditions. Chapter 10 in Solid-state NMR; applications in biomembrane structure. Edited by F. Separovic & M.-A. Sani; IOP Press (DOI 10.1088/978-0-7503-2532-5ch10)
[2] Li, M.; Mandal, A.; Tyurin, V. A.; DeLucia, M.; Ahn, J.; Kagan, V. E.; van der Wel, P. C. A. (2019) Surface-Binding to Cardiolipin Nanodomains Triggers Cytochrome c Pro-Apoptotic Peroxidase Activity via Localized Dynamics. Structure 2019, 27 (5), 806-815.e4. (DOI 10.1016/j.str.2019.02.007)
O-ChemS website available
The Solid-state NMR group is part of the O-ChemS consortium at the University of Groningen, working on supramolecular systems and materials that operate out of equilibrium. These materials borrow principles from life and nature, to attain unique functional properties. The website of this consortium is now live, available at: www.o-chems.nl.

Webinars of interest (during COVID)
During the COVID19 pandemic many conferences were cancelled, but also various interesting new online alternatives came into being. Below are a few relevant seminar series with relevance to our past and current research.
- Protein aggregation & chaperones
- NMR related
- Emerging Topics in Biomolecular Magnetic Resonance (YouTube)
- Global NMR Discussion meetings (YouTube)
- MIT SSNMR/DNP zoom seminars
- P3S seminars MLU Halle
- ICMRBS Early Careers Seminars
- Intercontinental NMR Seminar Series (ICONS2020)
- Membranes and lipids
- Biophysics
New publication on structure of mutant huntingtin protein from Huntington’s Disease
Congratulations to Dr. Jennifer Boatz and other team members for the acceptance and publication of a nice new paper on the structure of the misfolded mutant protein from Huntington’s disease. The paper is online at the Journal of Molecular Biology. Based on an integration of multiple techniques (NMR, EM, and X-ray diffraction), Jennifer assembled a new structural model of the protofilaments that make up the hierarchical fiber architecture of mutant huntingtin exon 1. This is a timely and important step forward in our understanding of the (mis)behaviour of the HD protein, and in particular how it forms pathogenic inclusions and protein aggregates.
In this new paper and prior work we (and others) have seen that the mutant protein is prone to form a collection of different kinds of aggregates, with each their own structure and functional properties. This is of substantial interest from a disease perspective, as these “functional properties” can encompass different degrees of neurotoxic properties. Surprisingly, Jennifer shows in this paper that one contributor to, or trigger of, huntingtin polymorphism is the concentration of the protein. Further studies will have to explore whether or how this finding impacts efforts to replicate cellular behaviour of the protein in vitro, and how it may affect the toxic properties of the aggregated protein states.
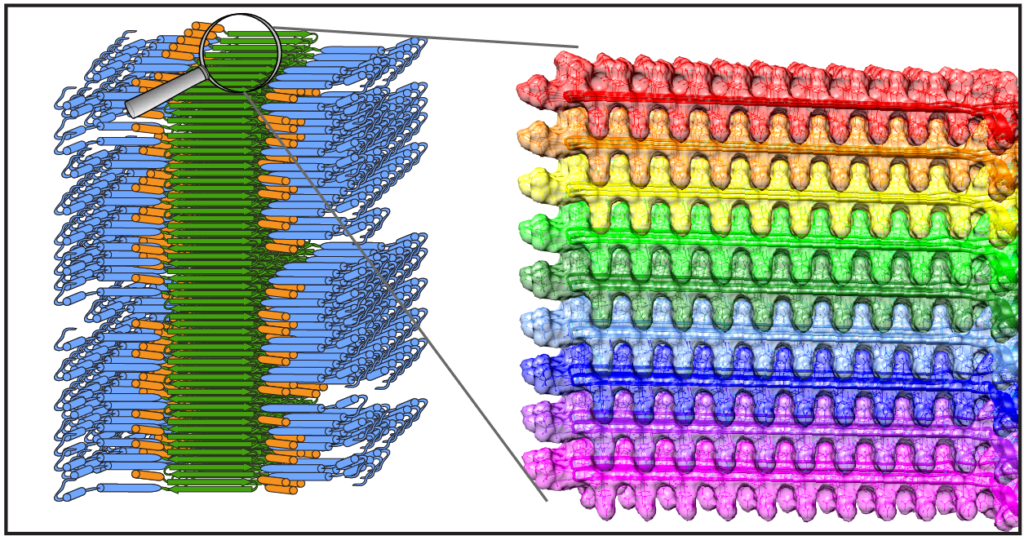
Reference:
Protofilament Structure and Supramolecular Polymorphism of Aggregated Mutant Huntingtin Exon 1. Boatz, J.C., Piretra, T., Lasorsa, A., Matlahov, I., Conway, J.F. & Van der Wel, P.C.A. (2020) J. Mol. Biol., 432(16): 4722-4744. [DOI] (Open Access)
Funding & support: The underlying research in this paper was enabled by funding support from the American NIH/NIGMS (grant R01 GM112678) and funding from the CampagneTeam Huntington in the Netherlands. For more information on the disease, see also our HD page and the CTH website. Other support came from the University of Groningen and the University of Pittsburgh.
Note added: Our Institute also highlighted this paper in a nice summary posted on the Zernike Institute website.
New review paper on ssNMR studies of polysaccharide hydrogels
PhD student Mustapha El Hariri El Nokab has put together a nice new review of solid-state NMR studies of polysaccharide hydrogels, which is now out in the journal Carbohydrate Polymers. This review is part of his research project that make use of solid-state NMR (and other tools) to look at functional polysaccharide hydrogels. This new research direction in the lab also constitutes part of our participation in the new Physics of Cancer (www.phycan.nl) initiative of the Zernike Institute for Advanced Materials.
Please find the open-access published paper at the journal Carbohydrate Polymers:
Competitive PhD Scholarships call open (deadline April 1st)
Our Institute and Faculty have opened up a competitive call for PhD scholarship applications. Our solid-state NMR group participates in one of the research theme areas, designated as “Advanced Materials”, which spans topics from physics of life, via bio-inspired materials, to materials and much more. The process is described in some detail on the RuG website. Briefly, applicants (with a MSc degree) are expected to contact a PI/supervisor (immediately!) and develop a fitting project to submit. The submission deadline of an initial idea (few hundred words) is due by April 1st 2020!
Suitable ideas are expected to fit within designated topics areas, described on the website here and here. Note especially also the 2nd link, as it contains important detailed information.T
Interested in this? Please contact us as soon as possible, to meet the tight deadlines. Together we can consider topics that range from self-assembling bio-inspired materials, Physics of Cancer, non-biological materials and other topics. Projects will be highly interdisciplinary as the involvement of a second supervisor is also required, with a distinct and complementary expertise.
New Publication on Lipid Oxidation
Now online in the journal Free Radical Biology and Medicine: a new review and perspective article by our collaborator Valerian Kagan (Univ. Pittsburgh). The paper (titled “Redox phospholipidomics of enzymatically generated oxygenated phospholipids as specific signals of programmed cell death”) examines the role of controlled lipid oxidation as a source of vital cellular signals. In particular it reviews recent work showing how cardiolipins and phosphatidylethanolamine lipids are oxidised by enzymes, as triggers of apoptosis and ferroptosis. Interestingly, the enzymatically generated oxidised species are distinct from those generated by spontaneous peroxidation, which may be important for the regulatory role of these species.
For more details, read the whole article here:
Kagan et al. (2020) “Redox phospholipidomics of enzymatically generated oxygenated phospholipids as specific signals of programmed cell death” Free Radical Biology and Medicine, Vol. 147, pp. 231-241
MAS ssNMR @ RuG arrived and operational (mostly)
Instrumentation update:
Our new 600 MHz NMR spectrometer from Bruker Biospin has arrived and been installed in building 5113 on the Zernike campus of the University of Groningen. The system features a new high-performance NEO-style console, with the necessary solid-state NMR (ssNMR) accessories. Although the ordered magic-angle-spinning (MAS) ssNMR probes are not yet available, a preliminary MAS ssNMR probe has been successfully installed.
For now, a few components are still missing, and the field remains a bit unstable. However, preliminary experiments have started! Further updates to follow.
For more information on our capabilities and for information about usage please contact the PI (p.c.a.van.der.wel@rug.nl) and/or our NMR technician (a.lasorsa@rug.nl).